Biological factors affecting prediction accuracy
Receptor gene expression
The quantile-normalized transcriptomes per cell type (level 3 data) will be used to determine whether the cognate receptor for each ligand in the previous experiment is present. Ligand-receptor interactions are from the Bader lab database included in CCInx.
## 91% (14%) of EGF receptors present in CMap data, either inferred or (assayed).
## 100% (8%) of HGF receptors present in CMap data, either inferred or (assayed).
## 94% (25%) of INS receptors present in CMap data, either inferred or (assayed).
## 100% (19%) of IGF2 receptors present in CMap data, either inferred or (assayed).
## 100% (33%) of TGFA receptors present in CMap data, either inferred or (assayed).
## 100% (15%) of IGF1 receptors present in CMap data, either inferred or (assayed).
## 94% (15%) of FGF1 receptors present in CMap data, either inferred or (assayed).
## 100% (31%) of HBEGF receptors present in CMap data, either inferred or (assayed).
## 97% (11%) of IL6 receptors present in CMap data, either inferred or (assayed).
## 98% (12%) of IL4 receptors present in CMap data, either inferred or (assayed).
## 93% (9%) of IFNG receptors present in CMap data, either inferred or (assayed).
## 100% (40%) of BTC receptors present in CMap data, either inferred or (assayed).
## 89% (11%) of GDNF receptors present in CMap data, either inferred or (assayed).
## 100% (0%) of GAS6 receptors present in CMap data, either inferred or (assayed).
## 99% (12%) of TNF receptors present in CMap data, either inferred or (assayed).
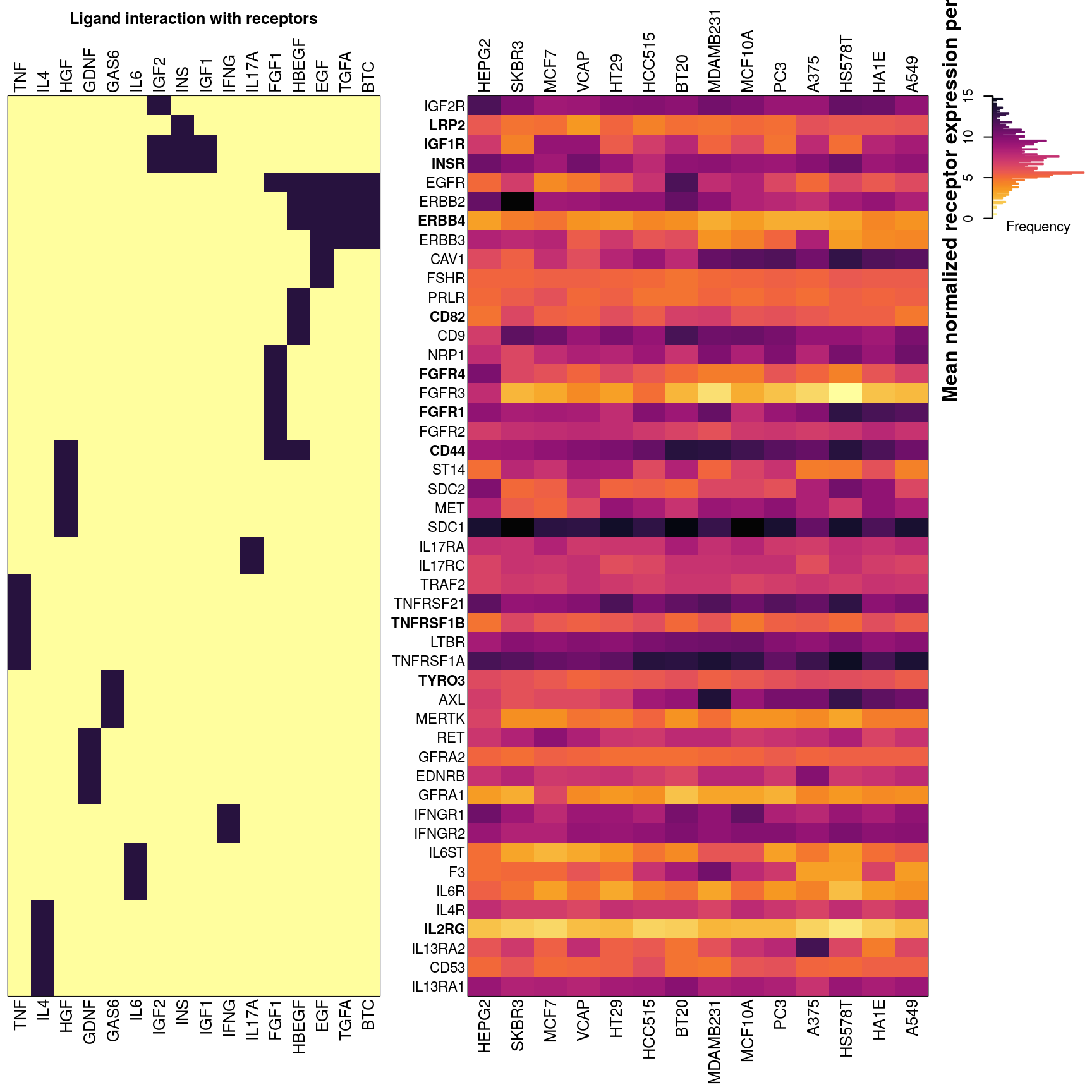
## 100% (9%) of IL17A receptors present in CMap data, either inferred or (assayed).Below is an overview of all potential cognate receptors per ligand present in the data. The left figure shows which ligands (dark purple) each receptor is known to interact with. On the right is a heatmap showing mean normalized gene expression per cell type for each receptor gene, with darker indicating higher expression. Gene names (rows) are ordered by heirarchical clustering of ligand-receptor interaction data, such that receptors common to the same ligand should be together. Landmark gene expression was emperically determined rather than inferred (names indicated in bold). Ligand names (columns, left) are ordered by heirarchical clustering of ligand-receptor interaction data, such that ligands sharing the same receptors should be proximal. Cell type names (columns, right) are ordered by heirarchical clustering of mean receptor gene expression.
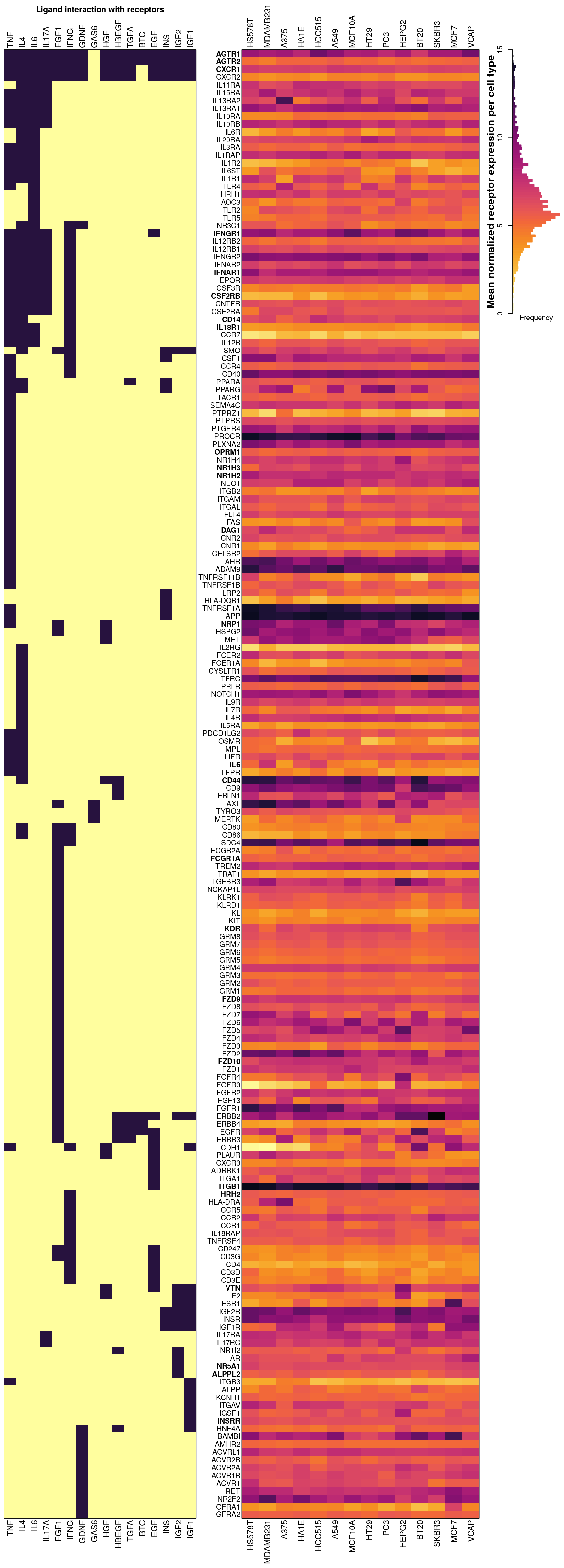
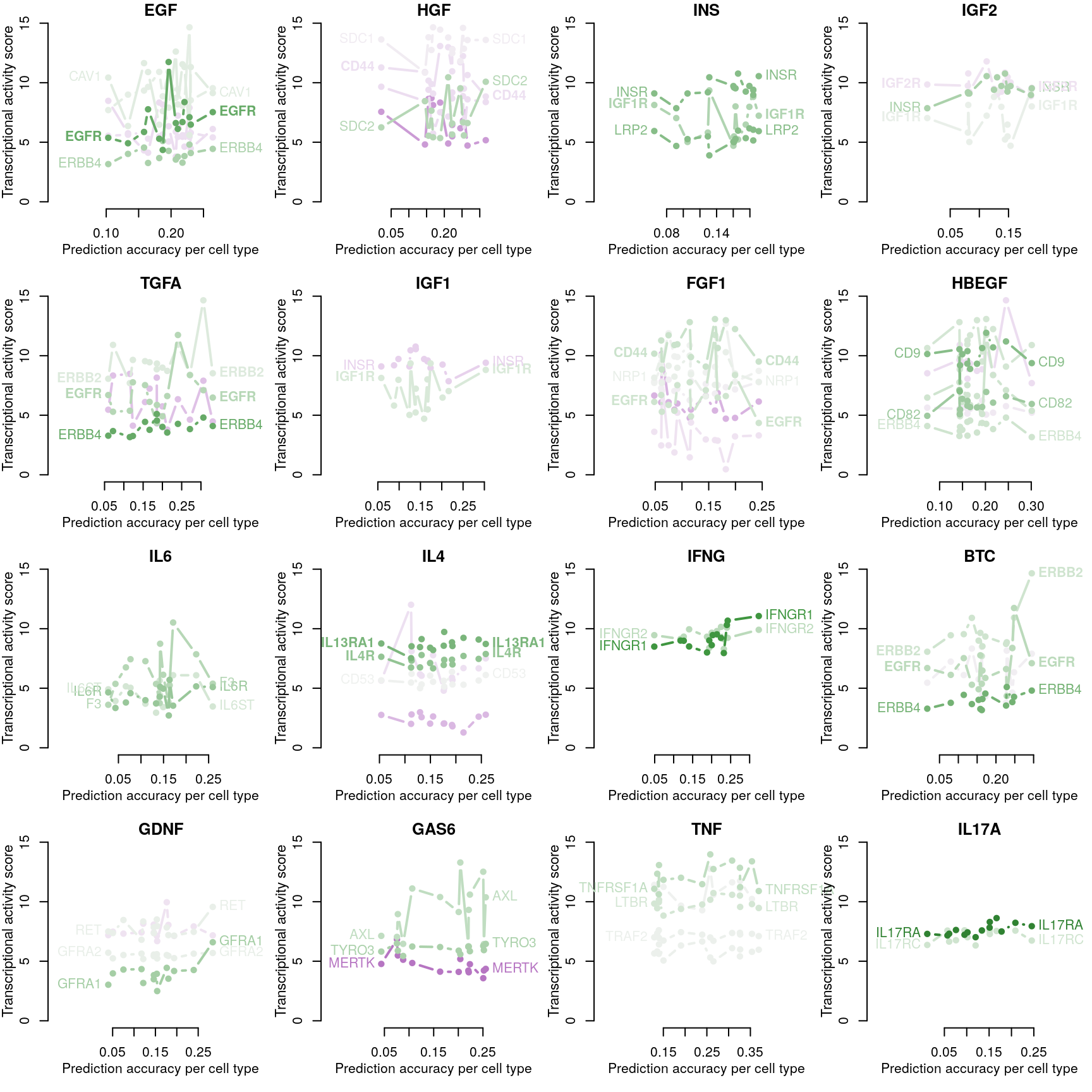
Receptor availability affecting ligand prediction accuracy
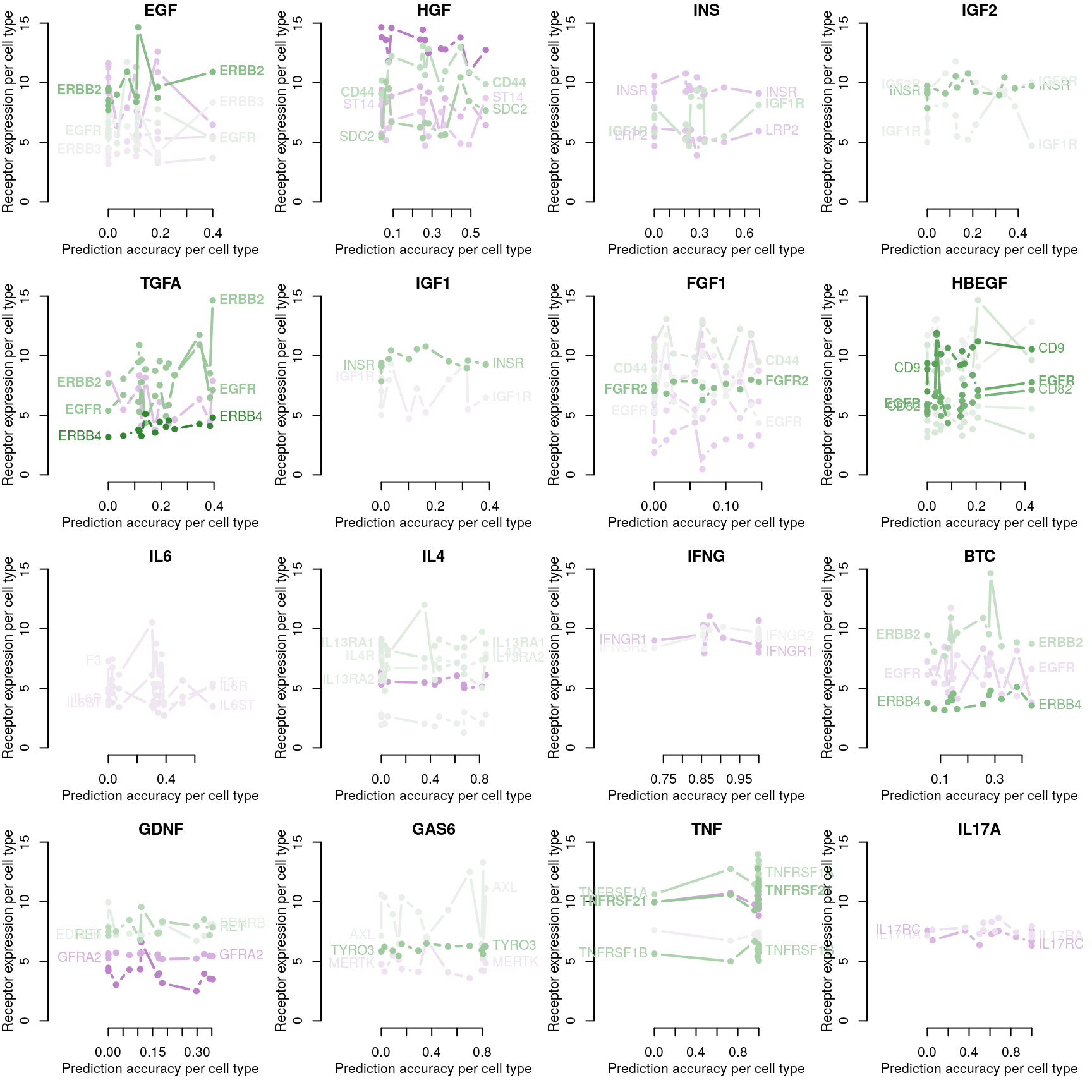
Cognate receptor expression for each ligand was compared to the accuracy per cell line of the leave-one-out ligand classification model outlined previously. Lines for each receptor are coloured by the spearman correlation coefficient between expression and accuracy per cell type (purple is negative, green is positive), and the three most correlated receptor genes are labeled.

Tumour necrosis factor alpha (TNF)
TNF binds to two receptors, TNFR1 and TNFR2, neither of which are present in the CMap data (neither assayed nor inferred). This is probably because they are (at least TNFR1) ubiquitously expressed, and thus their expression profiles are not informative or easily inferred. That may also explain why the transcriptional response to TNF treatment is so consistent - the may be receptor is present on all cell types. Downstream of TNF receptor activation, the multiple signaling pathways with conflicting phenotypes can be activated, though our data shows robust increases in expression of a common subset of genes across cell lines (see above).
With the Ramilowski et al L-R DB:
Receptor gene expression
The quantile-normalized transcriptomes per cell type (level 3 data) will be used to determine whether the cognate receptor for each ligand in the previous experiment is present. Ligand-receptor interactions are from the Bader lab database included in CCInx.
## NULL## 100% (50%) of EGF receptors present in CMap data, either inferred or (assayed).
## 100% (20%) of HGF receptors present in CMap data, either inferred or (assayed).
## 100% (33%) of INS receptors present in CMap data, either inferred or (assayed).
## 100% (67%) of IGF2 receptors present in CMap data, either inferred or (assayed).
## 100% (75%) of TGFA receptors present in CMap data, either inferred or (assayed).
## 100% (50%) of IGF1 receptors present in CMap data, either inferred or (assayed).
## 88% (50%) of FGF1 receptors present in CMap data, either inferred or (assayed).
## 100% (43%) of HBEGF receptors present in CMap data, either inferred or (assayed).
## 100% (0%) of IL6 receptors present in CMap data, either inferred or (assayed).
## 100% (40%) of IL4 receptors present in CMap data, either inferred or (assayed).
## 100% (0%) of IFNG receptors present in CMap data, either inferred or (assayed).
## 100% (75%) of BTC receptors present in CMap data, either inferred or (assayed).
## 80% (0%) of GDNF receptors present in CMap data, either inferred or (assayed).
## 100% (0%) of GAS6 receptors present in CMap data, either inferred or (assayed).
## 83% (17%) of TNF receptors present in CMap data, either inferred or (assayed).
## 100% (0%) of IL17A receptors present in CMap data, either inferred or (assayed).Below is an overview of all potential cognate receptors per ligand present in the data. The left figure shows which ligands (dark purple) each receptor is known to interact with. On the right is a heatmap showing mean normalized gene expression per cell type for each receptor gene, with darker indicating higher expression. Gene names (rows) are ordered by heirarchical clustering of ligand-receptor interaction data, such that receptors common to the same ligand should be together. Landmark gene expression was emperically determined rather than inferred (names indicated in bold). Ligand names (columns, left) are ordered by heirarchical clustering of ligand-receptor interaction data, such that ligands sharing the same receptors should be proximal. Cell type names (columns, right) are ordered by heirarchical clustering of mean receptor gene expression.
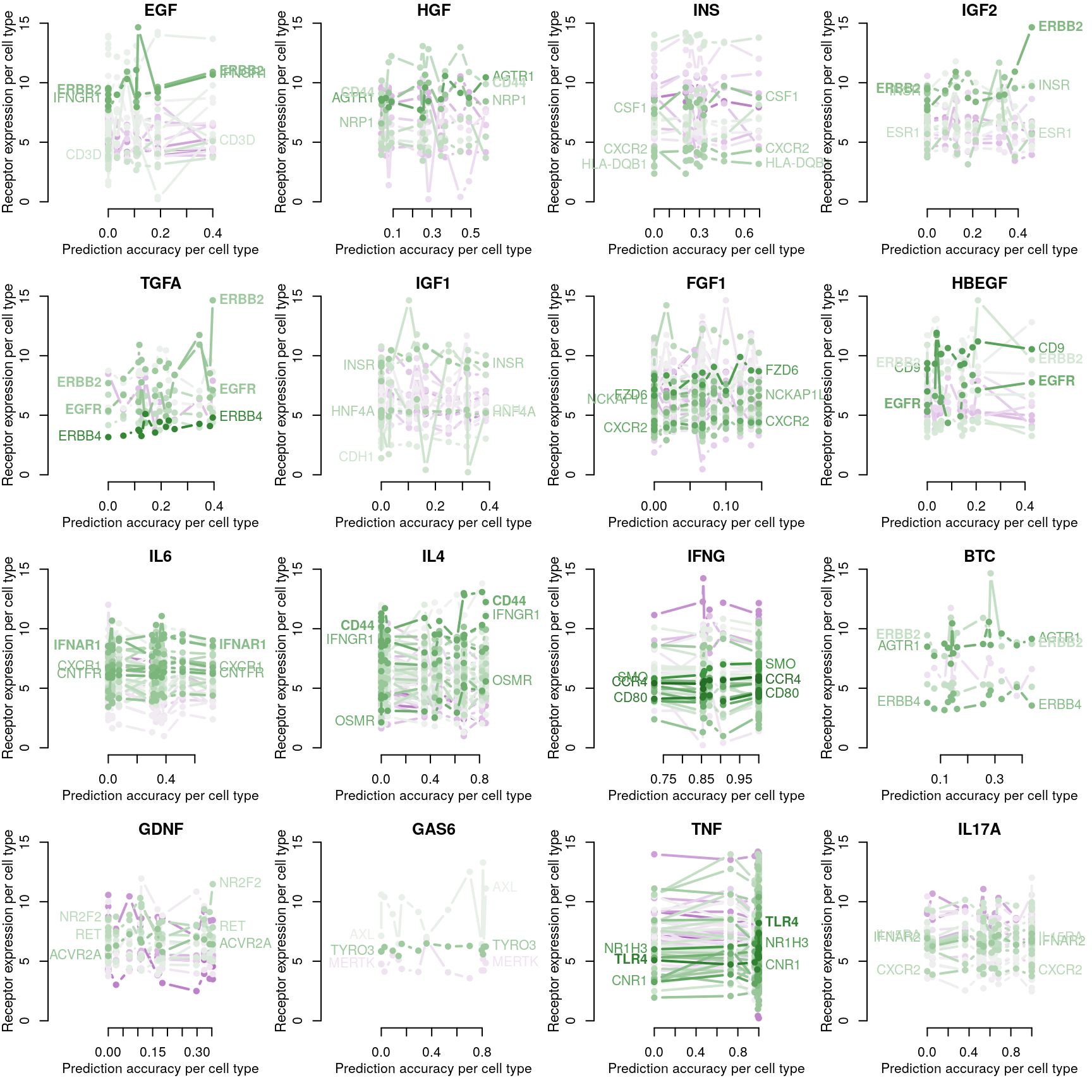
Receptor availability affecting ligand prediction accuracy
Cognate receptor expression for each ligand was compared to the accuracy per cell line of the leave-one-out ligand classification model outlined previously. Lines for each receptor are coloured by the spearman correlation coefficient between expression and accuracy per cell type (purple is negative, green is positive), and the three most correlated receptor genes are labeled.

Transcriptional Activity Score