Differentiating ligand-perturbed transcriptomes
by training on all cell types (Level 4 data)
Level 4 data
Since the level 4 data has replicates whereas the level 5 data collapses those into a single value, and training may perform better with more samples, here I repeat the positive control assay training a random forest classifier to identify selected ligand pertubands from transcriptomic changes across cell lines. The distribution of samples across ligands and cell lines is the same as the level 3 data, shown here.
Data saturation testing
Random forest model source code
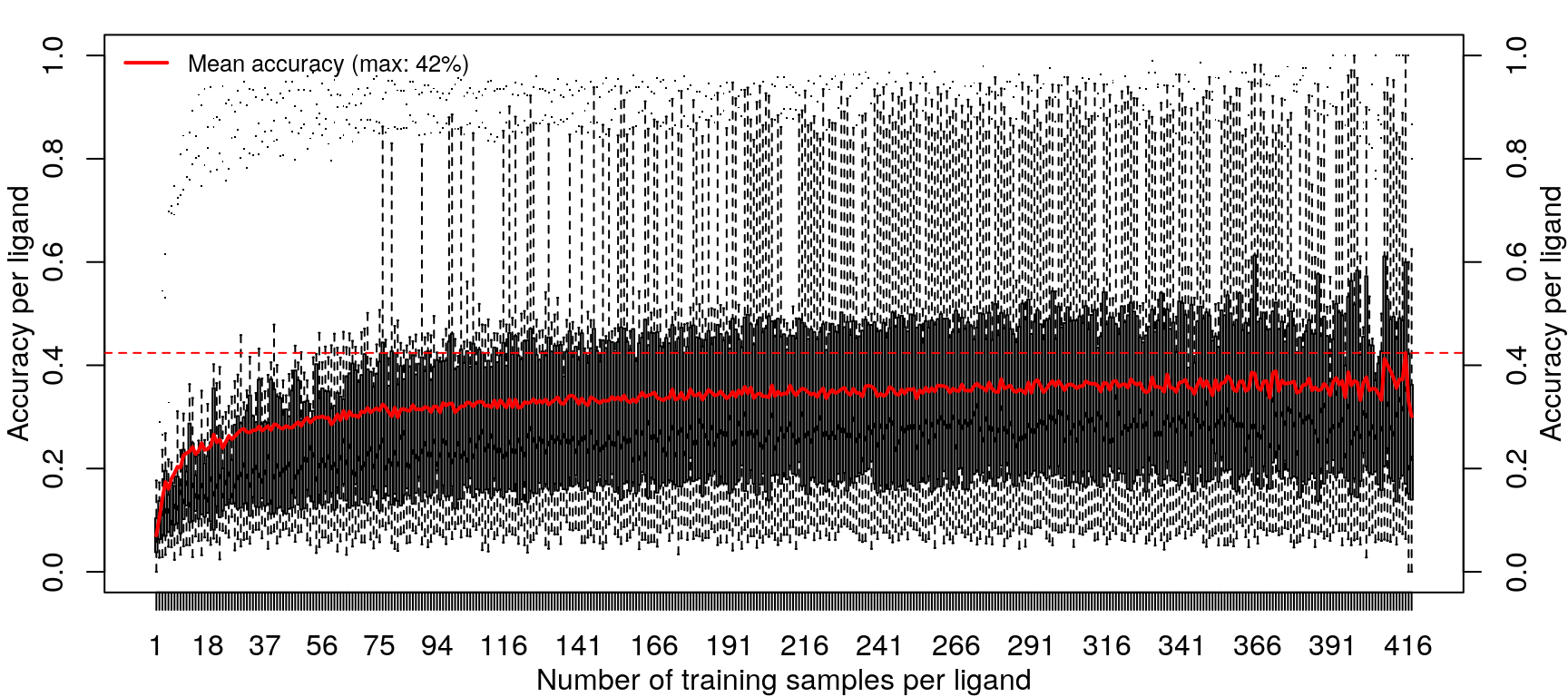
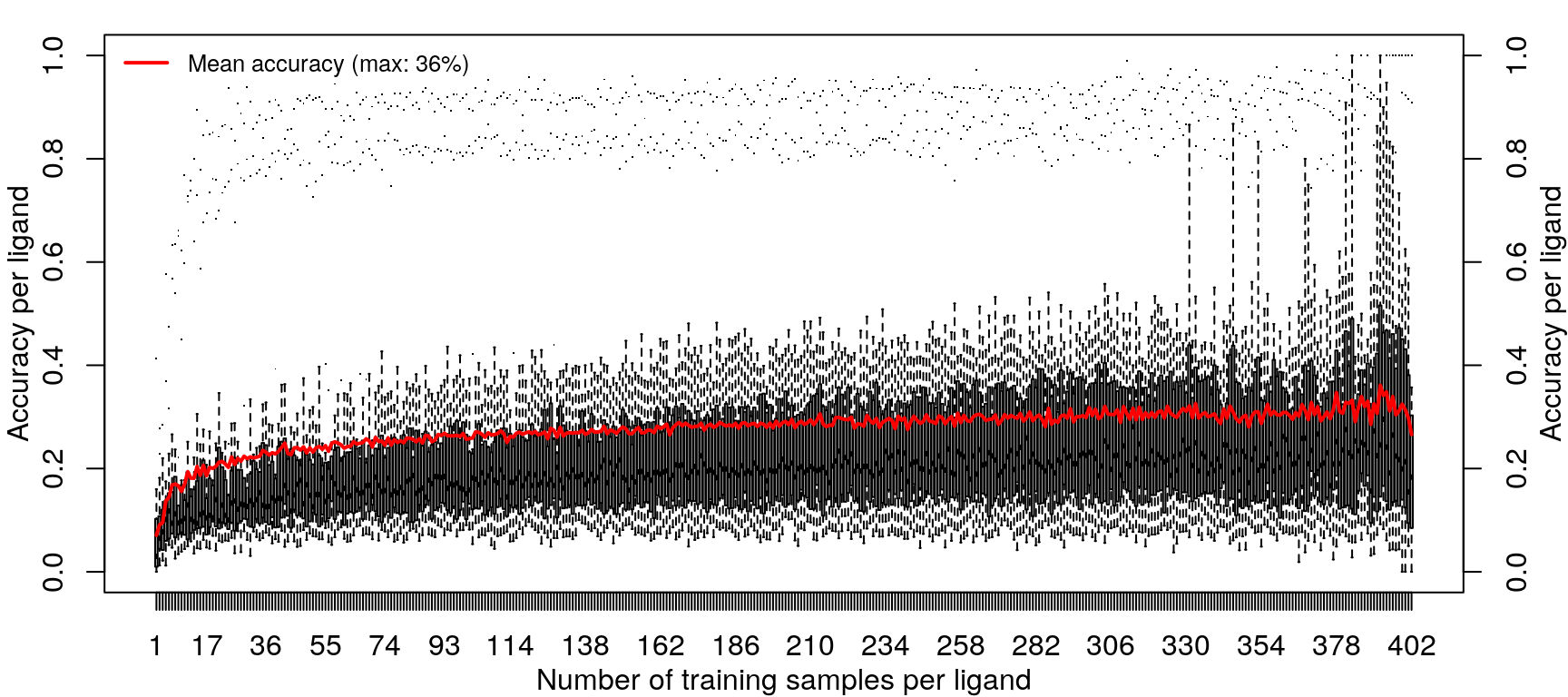
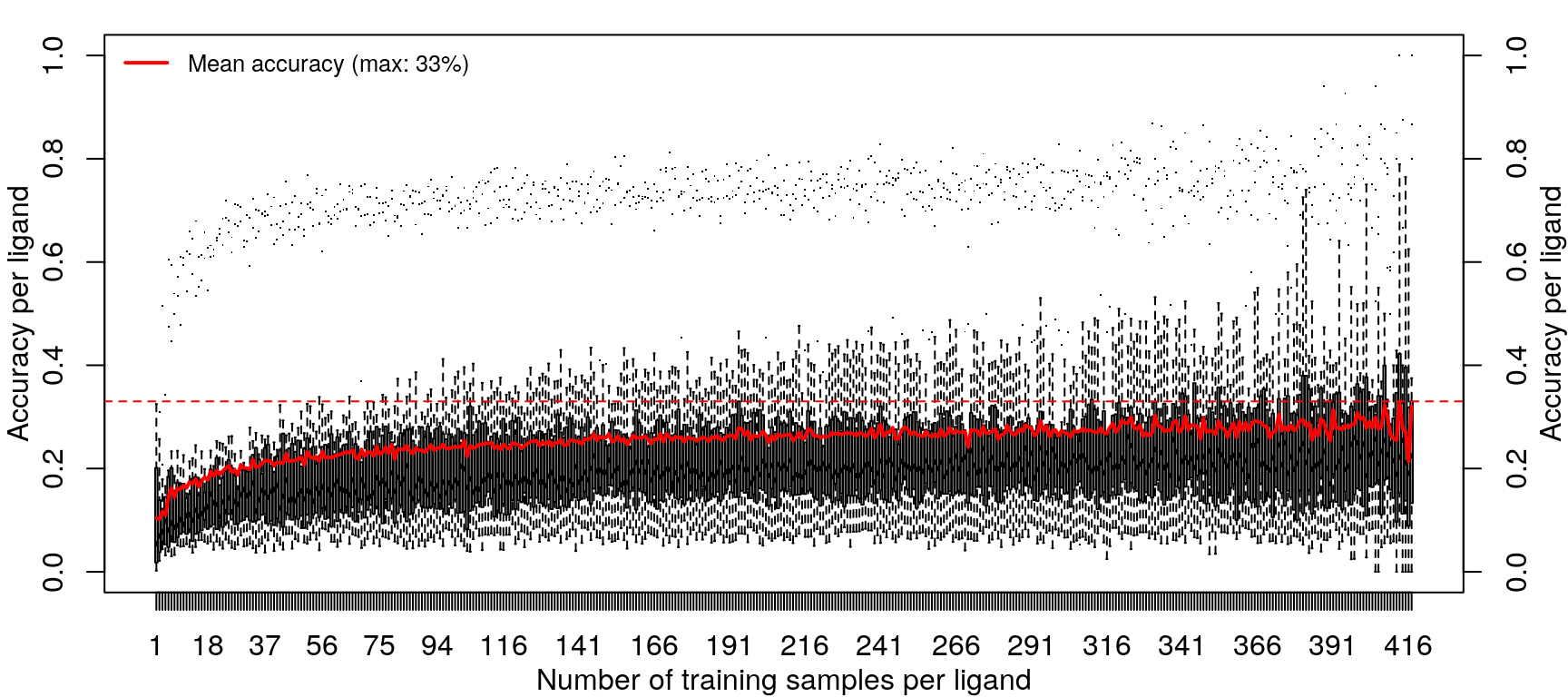
Training the random forest model was repeated with increasing data, from one sample per ligand (randomly sampled from the 14 cell types) to 418 samples per ligand (all but one sample for most ligands, except EGF).
Improvements in accuracy from increased training data level off after half the data is used, as was done in the previous models.
These results are not an improvement in overall accuracy compared to using the level 5 data (as seen here).
Improving prediction with metadata modelling
Random forest model source code
Adding metadata (cell type, treatment dosage and duration) as features in the model did not improve prediction accuracy in any appreciable manner.
Example sample distribution and results from the saturating test, at 300 training samples.
| BTC | EGF | FGF1 | GAS6 | GDNF | HBEGF | HGF | IFNG | IGF1 | IGF2 | IL17A | IL4 | IL6 | INS | TGFA | TNF | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| training | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 |
| testing | 131 | 524 | 138 | 126 | 128 | 138 | 175 | 132 | 139 | 140 | 118 | 133 | 135 | 140 | 139 | 122 |
| total | 431 | 824 | 438 | 426 | 428 | 438 | 475 | 432 | 439 | 440 | 418 | 433 | 435 | 440 | 439 | 422 |
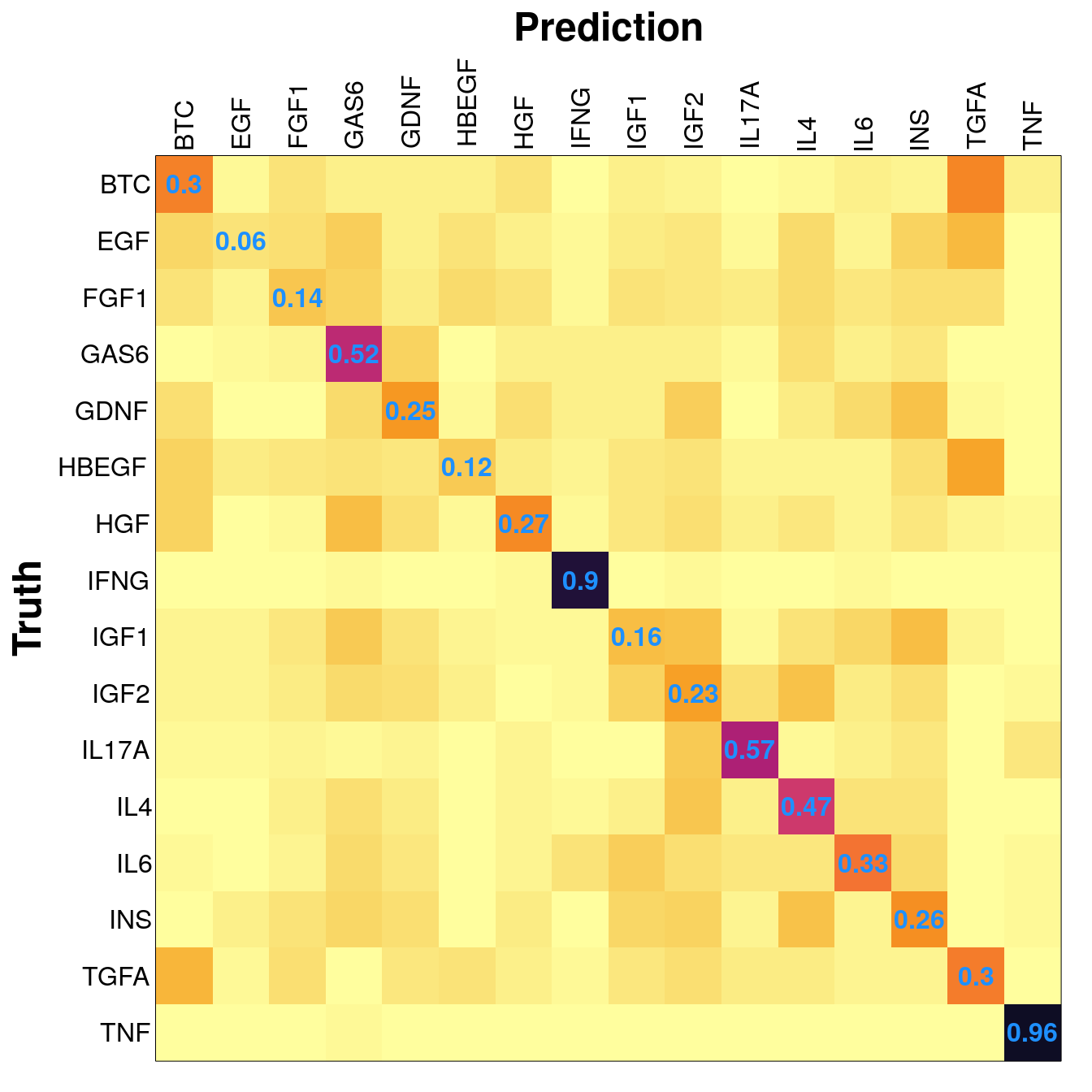
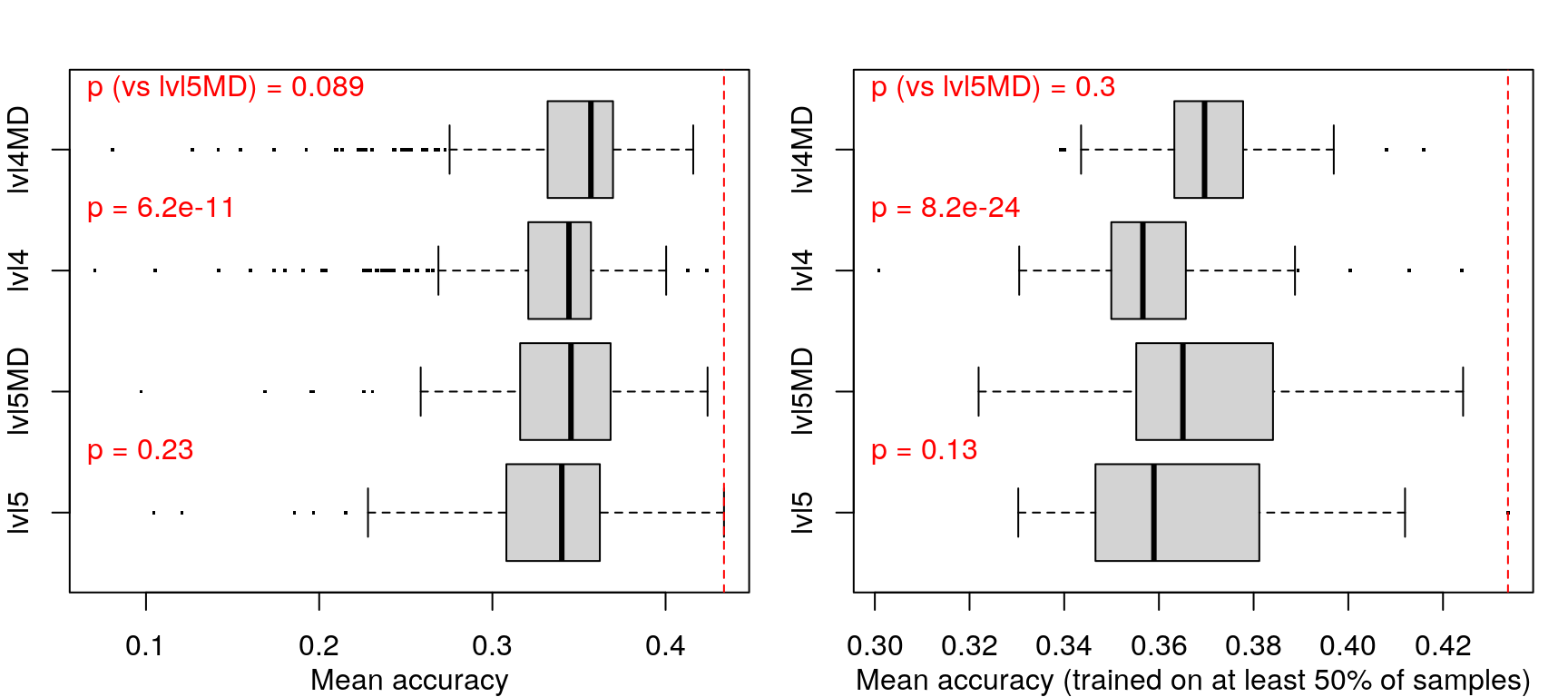
When trained on level 4 data (including metadata) less training samples were needed to achieve maximum accuracy, but maximum accuracy was pretty similar across the board (actually best in the first model).
Level 4 data recalculated
Data saturation testing
Random forest model source code
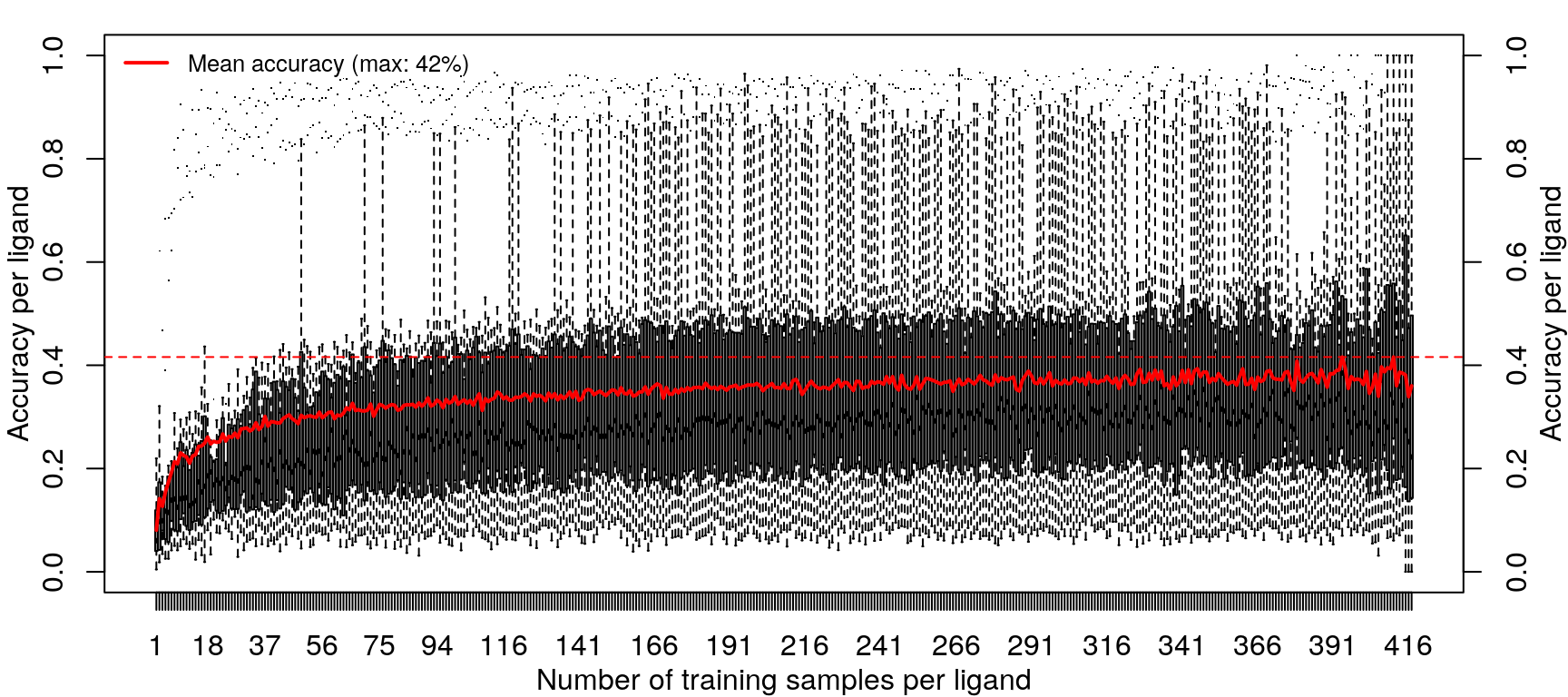
Training the random forest model was repeated with increasing data, from one sample per ligand (randomly sampled from the 14 cell types) to 418 samples per ligand (all but one sample for most ligands, except EGF).
Improvements in accuracy from increased training data level off after half the data is used, as was done in the previous models.
These results are not an improvement in overall accuracy compared to using the level 5 data (as seen here).
Improving prediction with metadata modelling
Random forest model source code
| BTC | EGF | FGF1 | GAS6 | GDNF | HBEGF | HGF | IFNG | IGF1 | IGF2 | IL17A | IL4 | IL6 | INS | TGFA | TNF | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| training | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 |
| testing | 113 | 505 | 119 | 108 | 110 | 119 | 137 | 113 | 121 | 121 | 118 | 115 | 116 | 121 | 120 | 103 |
| total | 413 | 805 | 419 | 408 | 410 | 419 | 437 | 413 | 421 | 421 | 418 | 415 | 416 | 421 | 420 | 403 |
Example distribution from the saturating test, at 300 training samples.
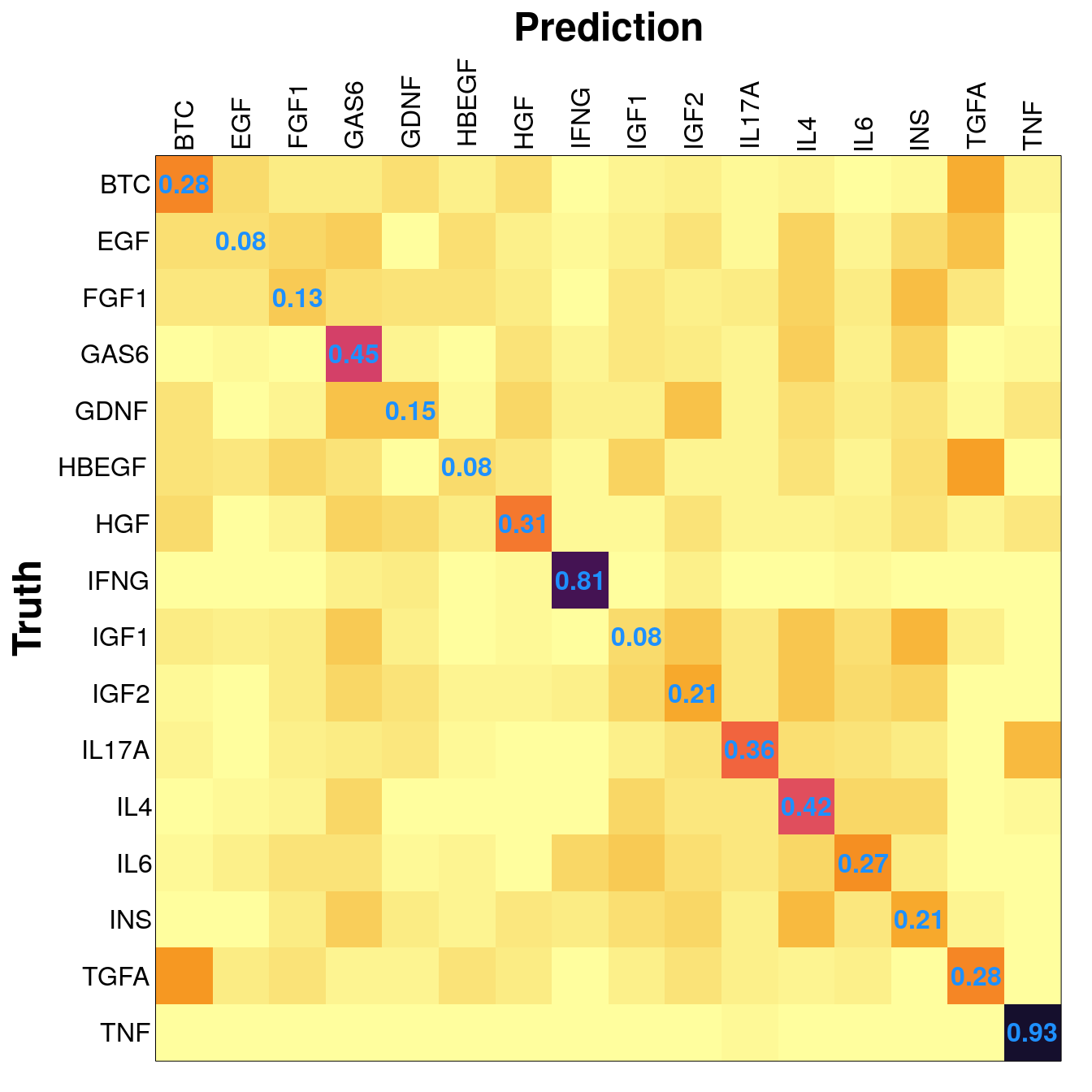
Adding metadata (cell type, treatment dosage and duration) as features in the model did not improve prediction accuracy in any appreciable manner.
## NULL## NULL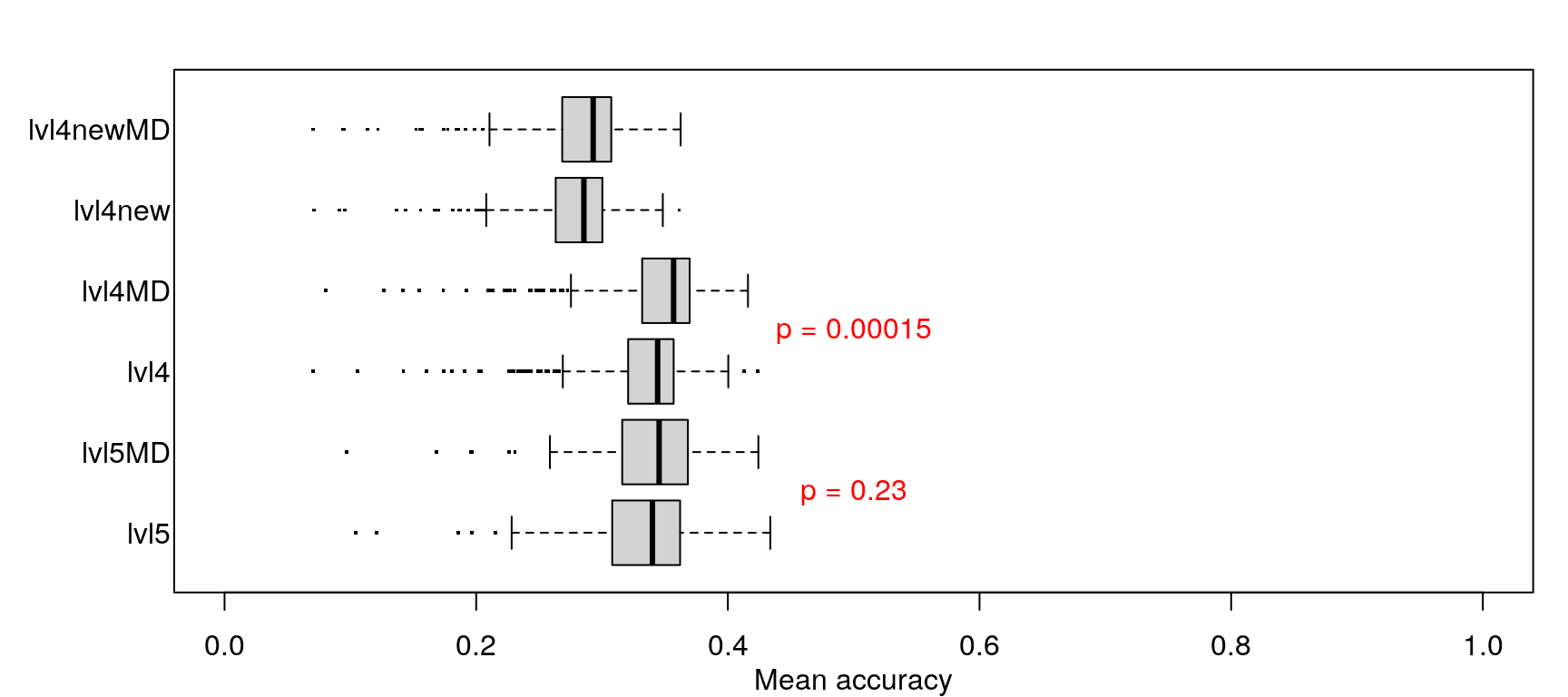
When trained on level 4 data (including metadata) less training samples were needed to achieve maximum accuracy, but maximum accuracy was pretty similar across the board (actually best in the first model).
PCA of level 4 data
Data saturation testing
First 100 PCs from the assayed Z-scores (level 4 data, ‘landmark’ genes only), with the addition of cell line, treatment dosage, and duration metadata, were used to train the random forest model.
Random forest model source code
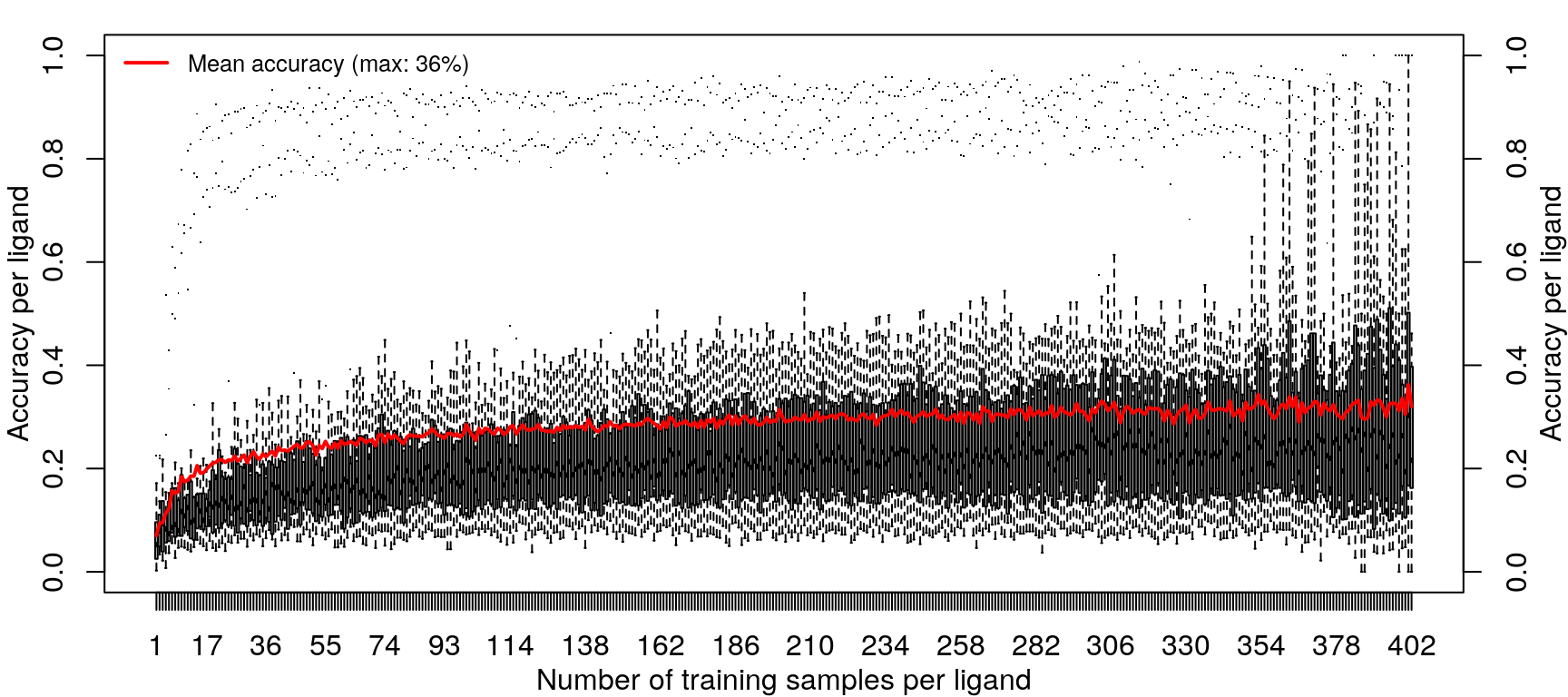
Training the random forest model was repeated with increasing data, from one sample per ligand (randomly sampled from the 14 cell types) to 418 samples per ligand (all but one sample for most ligands, except EGF).
Improvements in accuracy from increased training data level off after half the data is used, as was done in the previous models.
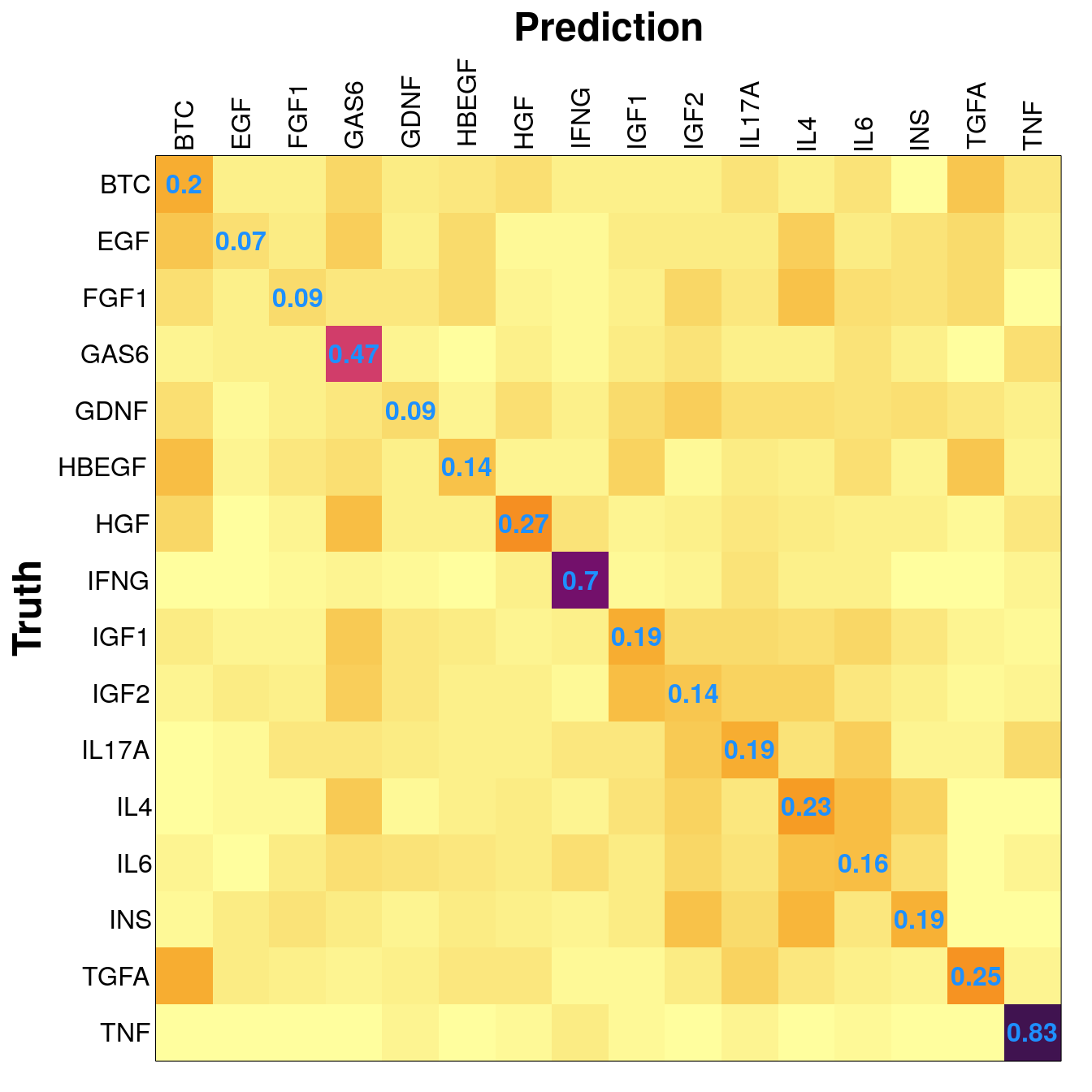
Example sample distribution and results from the saturating test, at 300 training samples.
| BTC | EGF | FGF1 | GAS6 | GDNF | HBEGF | HGF | IFNG | IGF1 | IGF2 | IL17A | IL4 | IL6 | INS | TGFA | TNF | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| training | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 | 300 |
| testing | 131 | 524 | 138 | 126 | 128 | 138 | 175 | 132 | 139 | 140 | 118 | 133 | 135 | 140 | 139 | 122 |
| total | 431 | 824 | 438 | 426 | 428 | 438 | 475 | 432 | 439 | 440 | 418 | 433 | 435 | 440 | 439 | 422 |
## NULL## NULL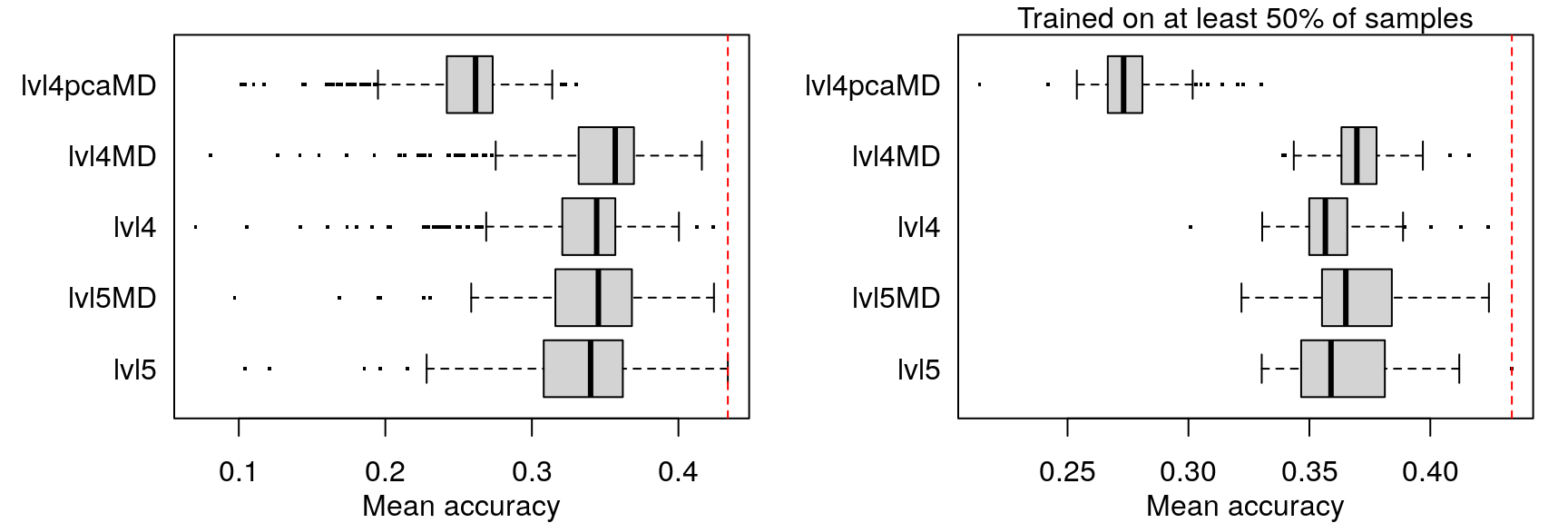
When trained on level 4 data (including metadata) less training samples were needed to achieve maximum accuracy, but maximum accuracy was pretty similar across the board (actually best in the first model).