Welcome
Welcome to Pathways and Network Analysis of -Omics Data 2024
Meet your Faculty
Gary Bader
Principal Investigator,
University of Toronto
 Dr. Bader develops biological network analysis and pathway information resources. He created the Biomolecular Interation Network Database ( BIND ) while working on his PhD and currently helps lead development of the free Cytoscape network visualization and analysis software Cytoscape.
Dr. Bader develops biological network analysis and pathway information resources. He created the Biomolecular Interation Network Database ( BIND ) while working on his PhD and currently helps lead development of the free Cytoscape network visualization and analysis software Cytoscape.
Lincoln Stein
Head, Adaptive Oncology,
OICR
 Dr. Stein played an integral role in many large-scale data initiatives including the development of the first physical clone map of the human genome, and running the data coordinating centre and the data portal for the SNP Consortium and the HapMap Consortium. Dr. Stein has also led the creation and development of Wormbase, a community model organism database for C. elegans, and Reactome, which is now the largest open community database of biological reactions and pathways. At OICR, Dr. Stein has led several international cancer data sharing and research initiatives, including the creation and development of the data coordination centre for the International Cancer Genome Consortium and other related projects. He continues to collaborate with national and international partners to create and promote data sharing standards, protocols and implementations.
Dr. Stein played an integral role in many large-scale data initiatives including the development of the first physical clone map of the human genome, and running the data coordinating centre and the data portal for the SNP Consortium and the HapMap Consortium. Dr. Stein has also led the creation and development of Wormbase, a community model organism database for C. elegans, and Reactome, which is now the largest open community database of biological reactions and pathways. At OICR, Dr. Stein has led several international cancer data sharing and research initiatives, including the creation and development of the data coordination centre for the International Cancer Genome Consortium and other related projects. He continues to collaborate with national and international partners to create and promote data sharing standards, protocols and implementations.
Gregory Schwartz
Scientist,
Princess Margaret Cancer Centre,
University Health Network
 Dr. Schwartz is a Scientist at the Princess Margaret Cancer Centre and Assistant Professor in the Department of Medical Biophysics at the University of Toronto. He has developed several methodologies for mutation detection, data integration, and cellular population visualization to understand cancer heterogeneity and diverse responses to anti-cancer therapies. His current research involves integrating multi-omic information and leveraging single-cell resolution to identify underlying mechanisms of drug resistance in cancer.
Dr. Schwartz is a Scientist at the Princess Margaret Cancer Centre and Assistant Professor in the Department of Medical Biophysics at the University of Toronto. He has developed several methodologies for mutation detection, data integration, and cellular population visualization to understand cancer heterogeneity and diverse responses to anti-cancer therapies. His current research involves integrating multi-omic information and leveraging single-cell resolution to identify underlying mechanisms of drug resistance in cancer.
Veronique Voisin
Research Associate,
Donnelly Centre for Cellular and Biomolecular Research,
University of Toronto
 Veronique is currently a bioinformatician applying pathway and networks analysis to high throughput genomics data for OICR cancer stem cell program. Previously, she worked on characterizing the gene signatures of different types of leukemias using a murine model
Veronique is currently a bioinformatician applying pathway and networks analysis to high throughput genomics data for OICR cancer stem cell program. Previously, she worked on characterizing the gene signatures of different types of leukemias using a murine model
Ruth Isserlin
Research data analyst,
Donnelly Centre for Cellular and Bimolecular Research,
University of Toronto
 Bioinformatician and data analyst in the Bader lab applying pathway and data analysis to varied data types. Developed Enrichment Map App for Cytoscape, an app to visually translate functional enrichment results from popular enrichment tools like GSEA to networks. Further developed the Enrichment Map Pipeline including development of additional Apps to help summarize and analyze resulting Enrichment Maps, including PostAnalysis, WordCloud, and AutoAnnotate App.
Bioinformatician and data analyst in the Bader lab applying pathway and data analysis to varied data types. Developed Enrichment Map App for Cytoscape, an app to visually translate functional enrichment results from popular enrichment tools like GSEA to networks. Further developed the Enrichment Map Pipeline including development of additional Apps to help summarize and analyze resulting Enrichment Maps, including PostAnalysis, WordCloud, and AutoAnnotate App.
Chaitra Sarathy, PhD
Bioinformatics Specialist,
Krembil Research Institute,
University Health Network
 Dr. Sarathy is a computational biologist with industry experience in software development. Her previous research focussed on developing multi-scale mathematical models of human systems to characterise biochemical changes in obesity. In addition, she has developed methods based on machine learning and multi-omics integration to identify drug targets in cancer and stratify patients for clinical trials. She currently focusses on characterising genetic malfunctions in neurological diseases.
Dr. Sarathy is a computational biologist with industry experience in software development. Her previous research focussed on developing multi-scale mathematical models of human systems to characterise biochemical changes in obesity. In addition, she has developed methods based on machine learning and multi-omics integration to identify drug targets in cancer and stratify patients for clinical trials. She currently focusses on characterising genetic malfunctions in neurological diseases.
Nia Hughes
Program Manager, Bioinformatics.ca
Toronto, ON, CA
nia.hughes@oicr.on.ca
 Nia is the Program Manager for Bioinformatics.ca, where she coordinates the Canadian Bioinformatics Workshop Series. Prior to starting at the OICR, she completed her M.Sc in Bioinformatics from the University of Guelph in 2020 before working there as a bioinformatician studying epigenetic and transcriptomic patterns across maize varieties.
Nia is the Program Manager for Bioinformatics.ca, where she coordinates the Canadian Bioinformatics Workshop Series. Prior to starting at the OICR, she completed her M.Sc in Bioinformatics from the University of Guelph in 2020 before working there as a bioinformatician studying epigenetic and transcriptomic patterns across maize varieties.
Thank you for attending the Pathway and Network Analysis of Omics Data workshop! Help us make this workshop better by filling out our survey.
Class Materials
You can download the printed course manual here.
Pre-Workshop Materials and Laptop Setup Instructions
Laptop Setup Instructions
A Check list to setup your laptop can be found here
Install these tools on your laptop before coming to the workshop:
Basic programs
- A robust text editor:
- For Windows/PC - notepad++
- For Linux - gEdit
- For Mac – TextWrangler
- For Windows/PC - notepad++
- A file decompression tool.
- A robust internet browser such as:
- Firefox
- Safari
- Chrome
- Microsoft Edge
- A PDF Viewer
- Adobe Acrobat or equivalent
Cytoscape Installation
Please install the latest version of Cytoscape 3.10.2 or Cytoscape Download as well as a group of Cytoscape Apps that we will be using for different parts of the course.
- Install Cytoscape 3.10.2:
- Go to: https://github.com/cytoscape/cytoscape/releases/3.10.2/ OR https://cytoscape.org/download.html
- Choose the version corresponding to your operating system (OS, Windows or UNIX)
- Follow instructions to install cytoscape
- Verify that Cytoscape has been installed correctly by launching the newly installed application
- Install the following Cytoscape Apps - Apps are installed from within Cytoscape.
- In order to install Apps launch Cytoscape
- From the menu bar, select ‘Apps’, then ‘App Store’, then ‘Show App Store’.
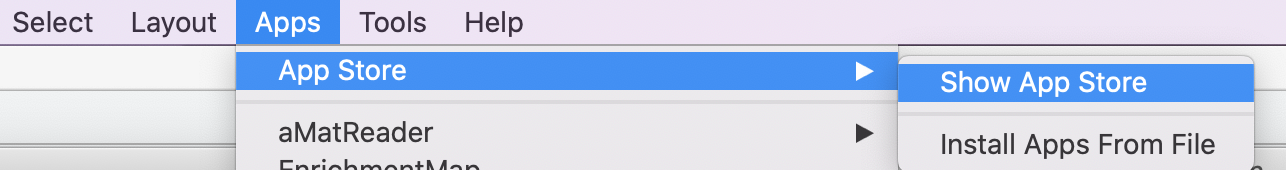
- App Store will appear in left hand Panel

- Within search bar at the top of the panel, search for the app listed below. Once you click on search icon a web browser will be launched with the apps that match your search.
- Select the correct app (there might be a few that match your search term).
- Click on “Install”
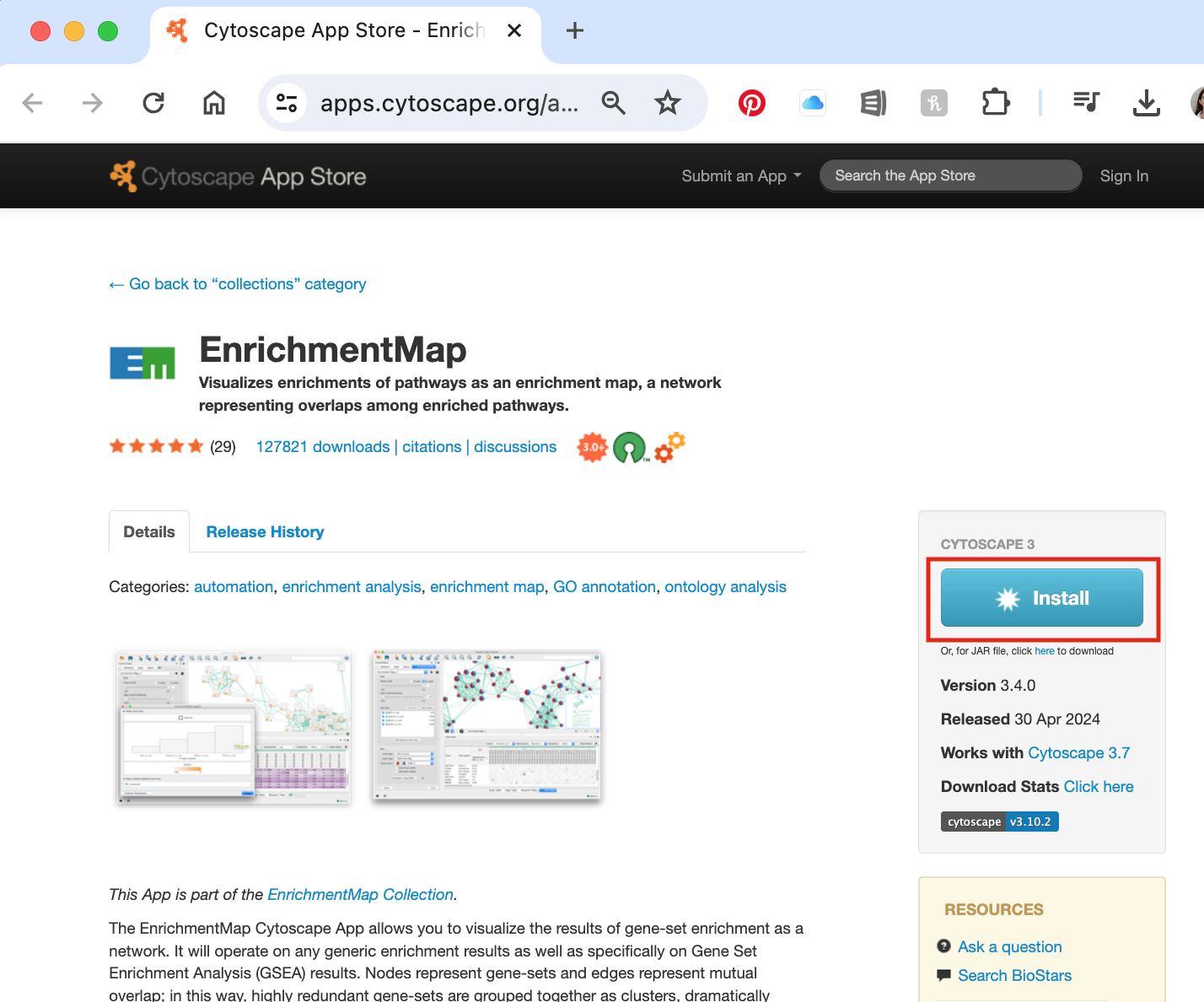
- install the following:
- EnrichmentMap 3.4.0
- EnrichmentMap Pipeline Collection 1.1.0 (it will install ClusterMaker2 v2.3.4, WordCloud v3.1.4 and AutoAnnotate v1.5.0)
- GeneMANIA 3.5.3
- IRegulon 1.3
- ReactomeFIPlugin 8.0.6 - http://apps.cytoscape.org/apps/reactomefiplugin
- stringApp 2.0.3
- scNetViz 1.7.1
- yFiles Layout Algorithms 1.1.4
- In order to install Apps launch Cytoscape
- Install the data set within GeneMANIA app. This requires time and a good network connection to download completely (~15mins)
- From the menu bar, select ‘Apps’, hover over ‘GeneMANIA’, then select ‘Choose Another Data Set’.
- From the list of available data sets, select the most recent and under ‘Include only these networks:’ select ‘all’. Click on ‘Download’.
- An ‘Install Data’ window will pop-up. Select H.Sapiens Human (2589 MB). Click on ‘Install’.
GSEA Installation
Please install the latest version of GSEA (4.3.3)
- Download GSEA
- Go to the GSEA page
- Register (using an institutional email address)
- Login
- Locate the Download page and download the version corresponding to your system
- MAC users: download GSEA_4.3.3.app.zip
- Window users: download GSEA_Win_4.3.3-installer.exe
- Unix users: download GSEA_Linux_4.3.3.zip
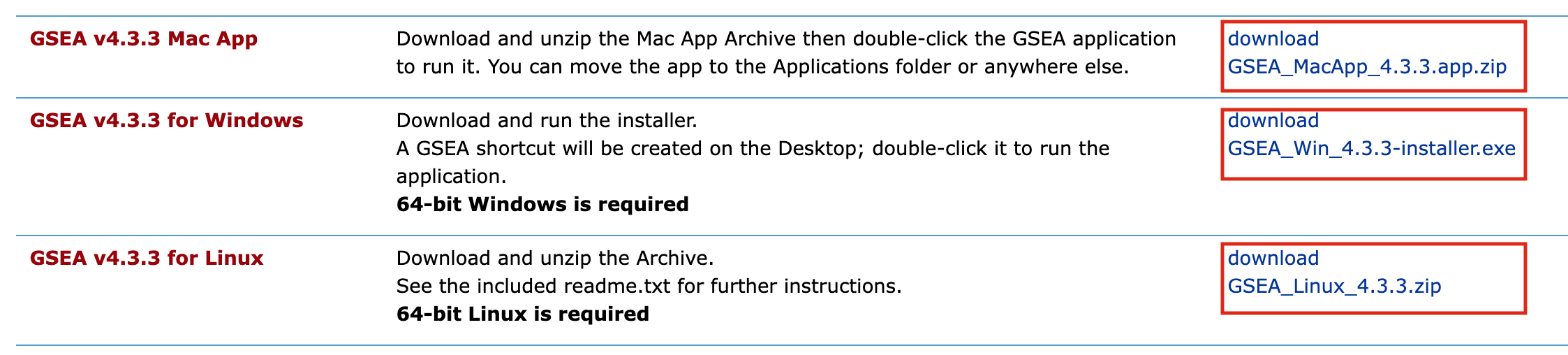
- Launch GSEA to test it.
- Go to the GSEA page
- Download GSEA for command line : this is necessary for all platform users to run GSEA from a script (integrated workflow on day 3)
- Download GSEA_4.3.3.zip (and keep it for later use during the workshop)

Docker Installation
Docker is a virtualization software that allows you to run programs isolated from your current laptop set up. It eases the burden of installing multiple software requirements and packages.
- Please install the latest version of Docker Desktop.
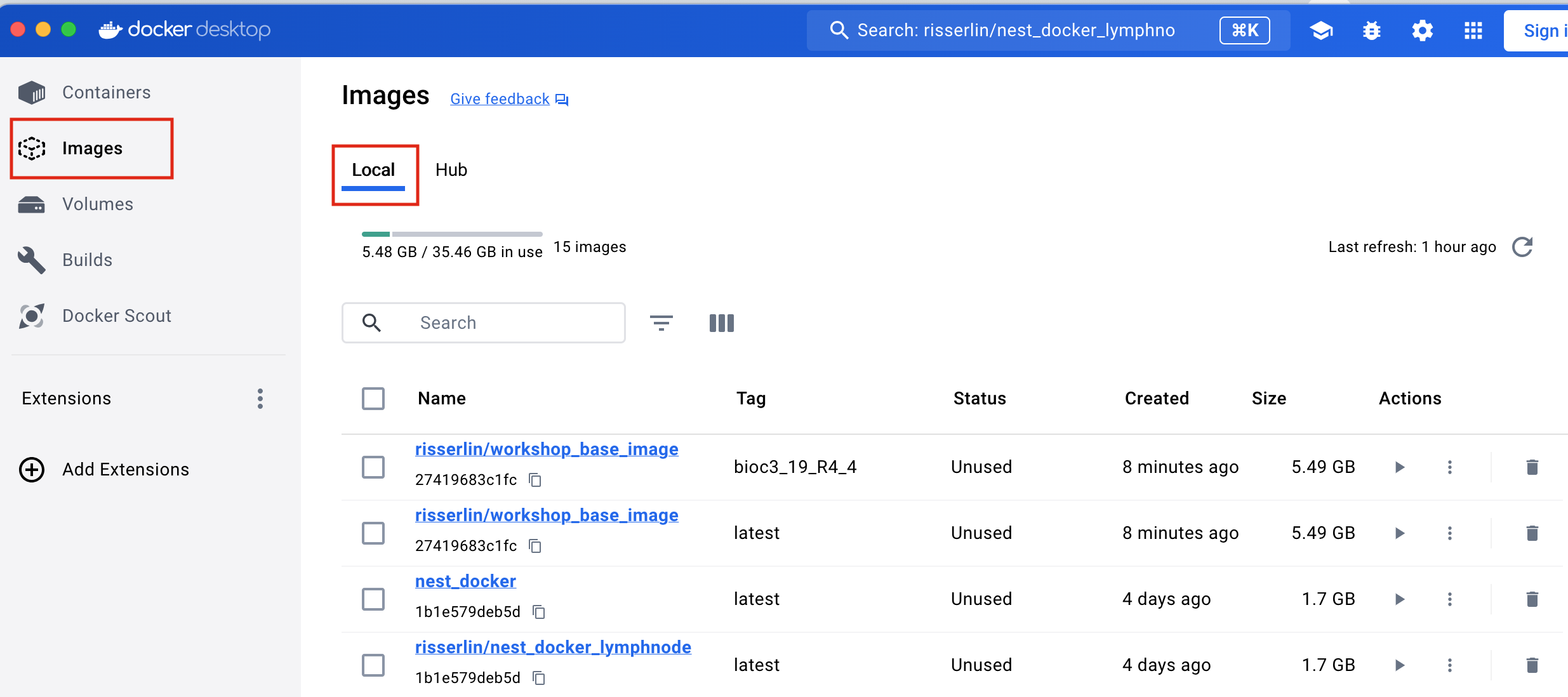
- Pull the required images used in the course
- Open docker desktop (If docker is already running you can find the docker icon in your task bar. Right click on the icon and select “Go to Dashboard”)
- Find the search bar in the docker desktop dashboard
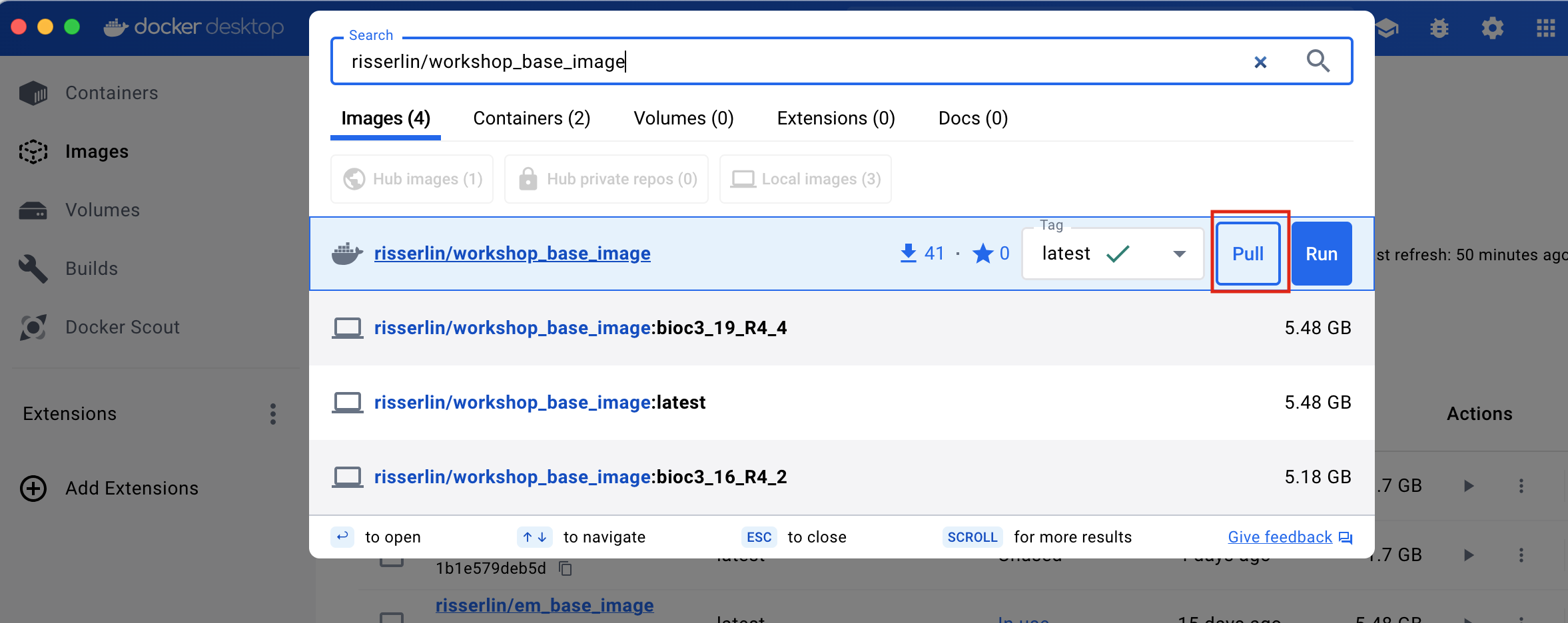
- Enter “risserlin/workshop_base_image” into the search bar at the top of the docker desktop dashboard.
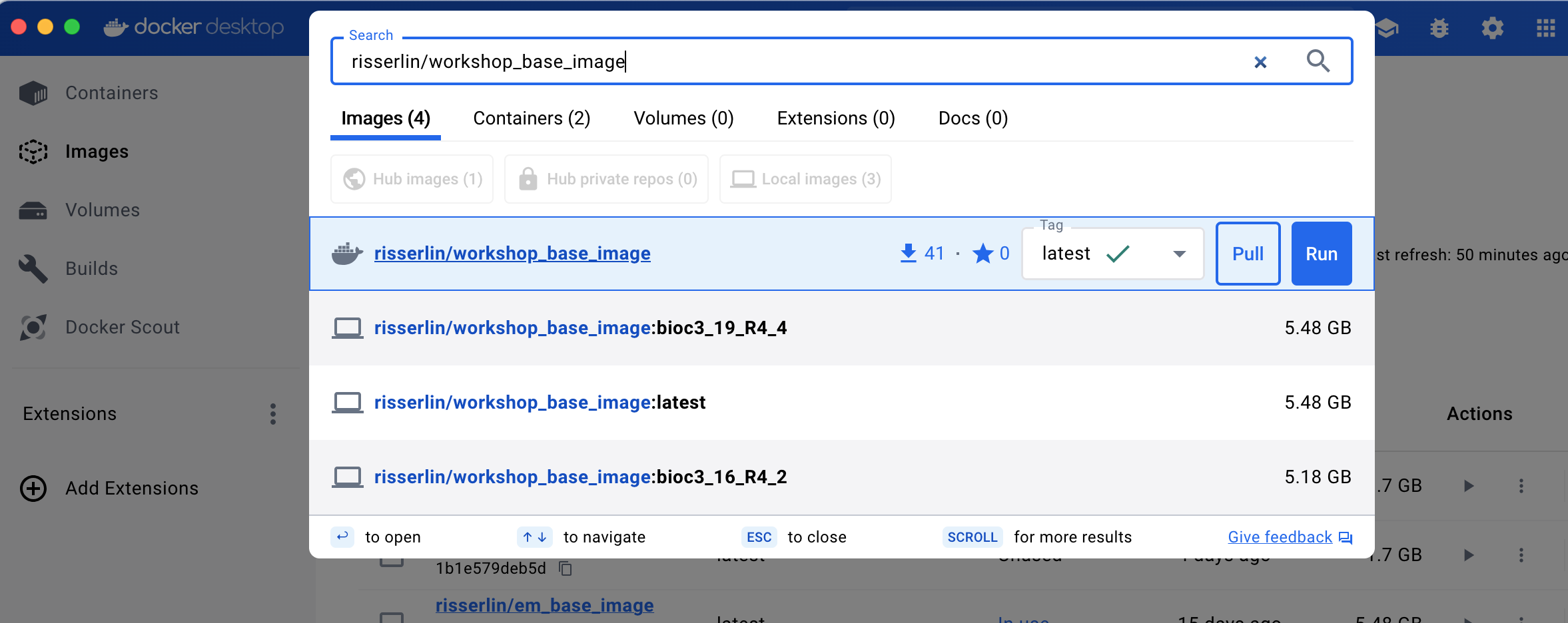
- Click on “Pull” to download the image.
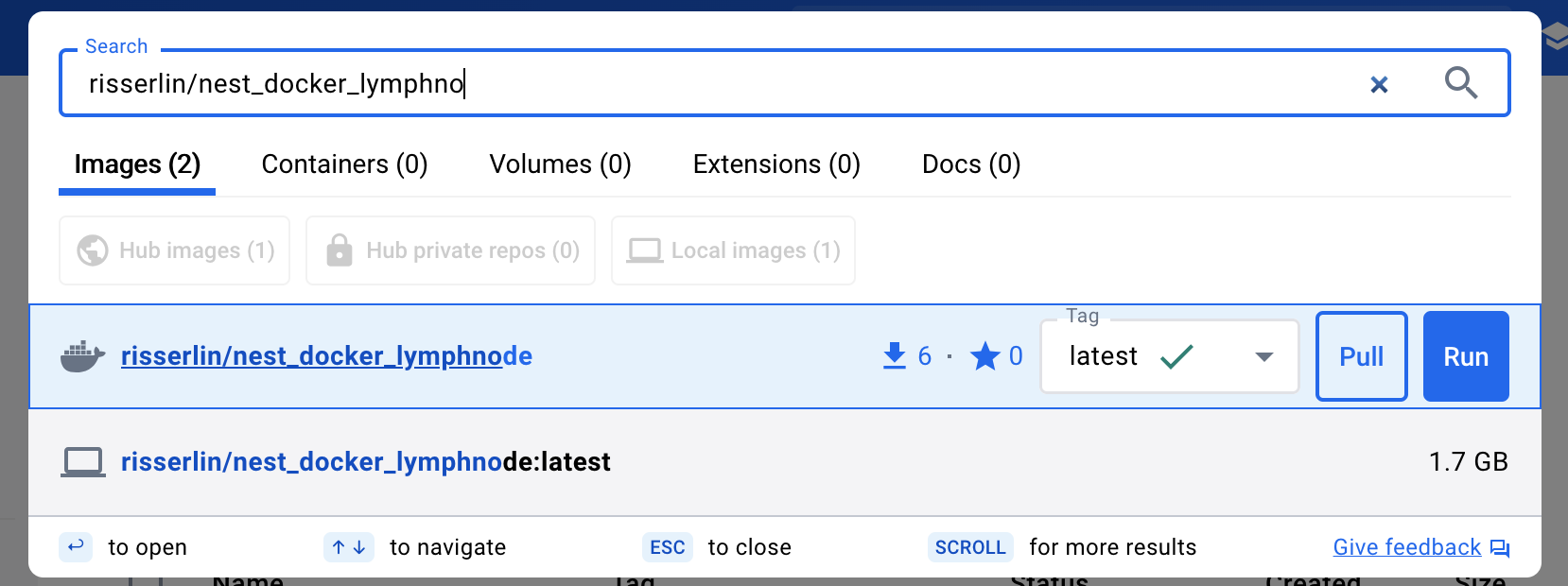
- Enter “risserlin/nest_docker_lymphnode” into the search bar at the top of the docker desktop dashboard.
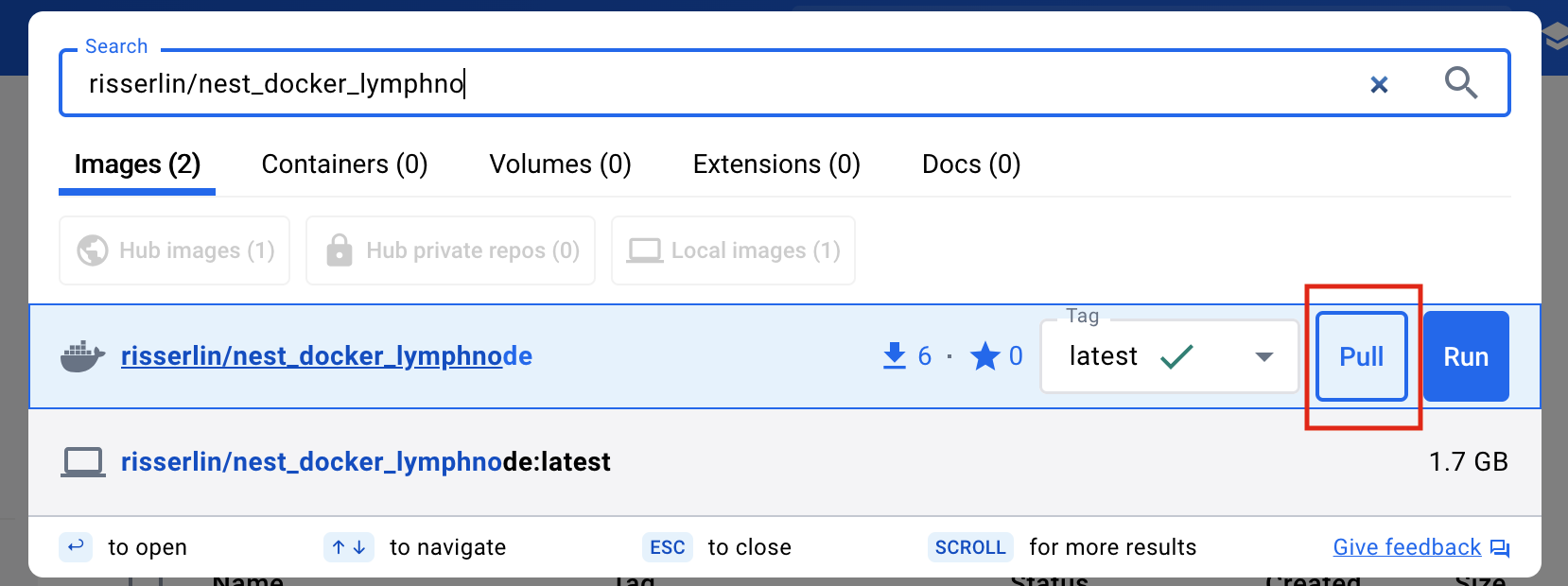
- Click on “Pull” to download the image.
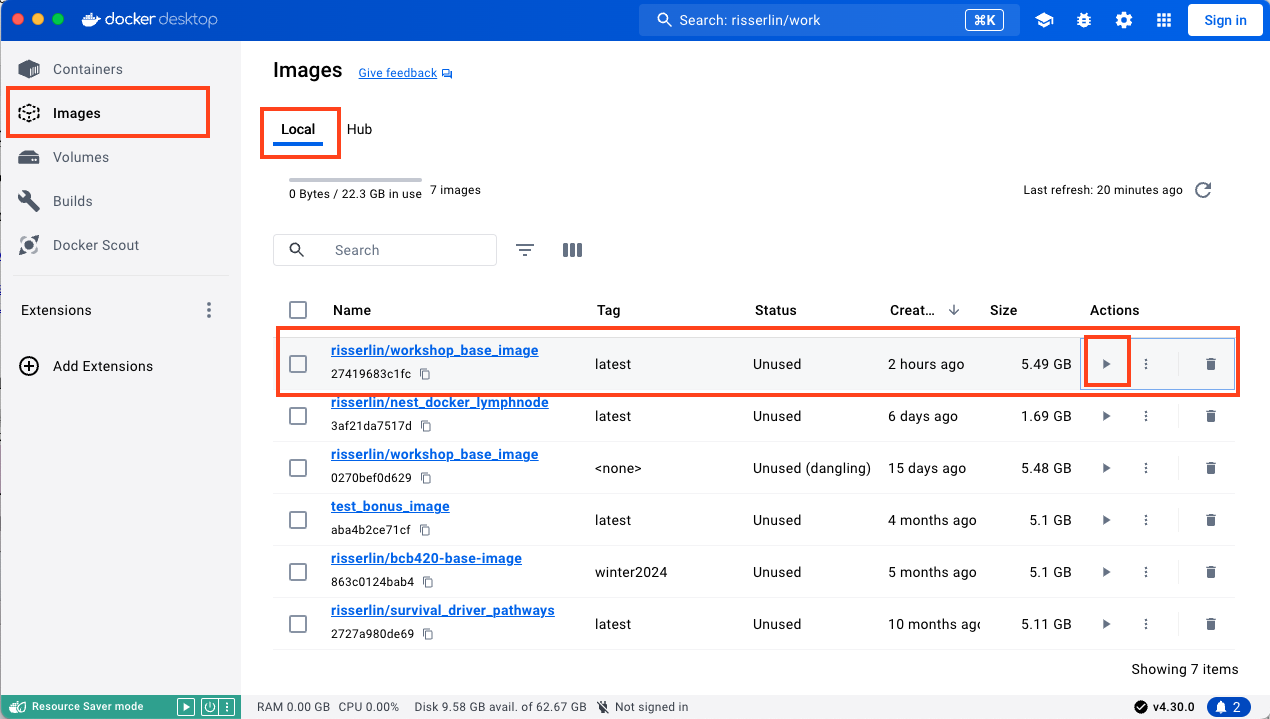
- You should now see both of your images listed in the docker desktop image section (in the local tab)
Pre-workshop Tutorials
It is in your best interest to complete these before the workshop.
Cytoscape Preparation tutorials
Go to : https://github.com/cytoscape/cytoscape-tutorials/wiki and follow :
R Tutorial
Use your newly installed docker workshop_base_image to try out R and go through the following tutorial -
- R tutorial - There will be instructions on how to install R and RStudio in the tutorial. Instead of installing use the workshop_base_image docker image that you installed above as follows:
- Open docker desktop (If docker is already running you can find the docker icon in your task bar. Right click on the icon and select “Go to Dashboard”)

- Click on Images –> Local –> And find the workshop_base_image. click on the Play button
- Expand the ‘optional settings’
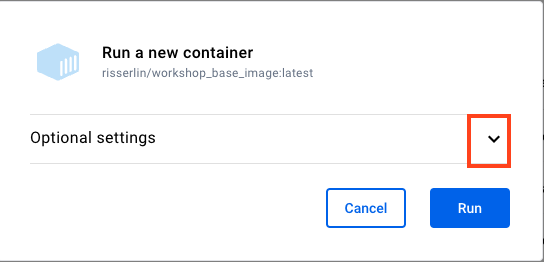
- Change -
- ‘container name’ to R_tutorial,
- ‘Host Port’ to 8787,
- Add environment variable PASSWORD and set value to password
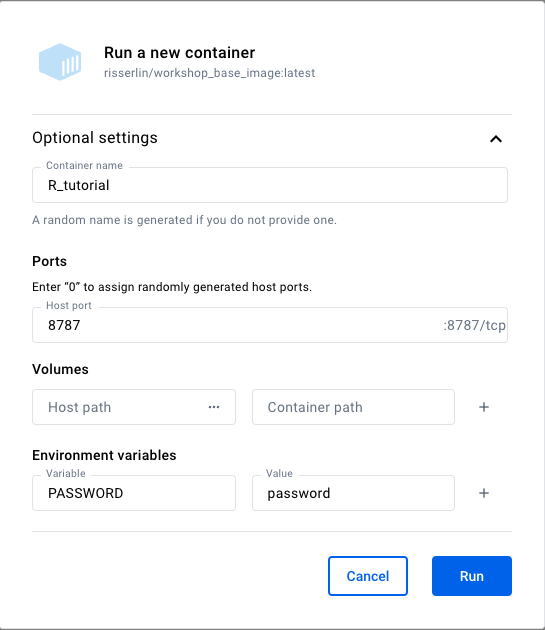
- Click on ‘Run’. Docker will display a tab with all the information about the container you just launched
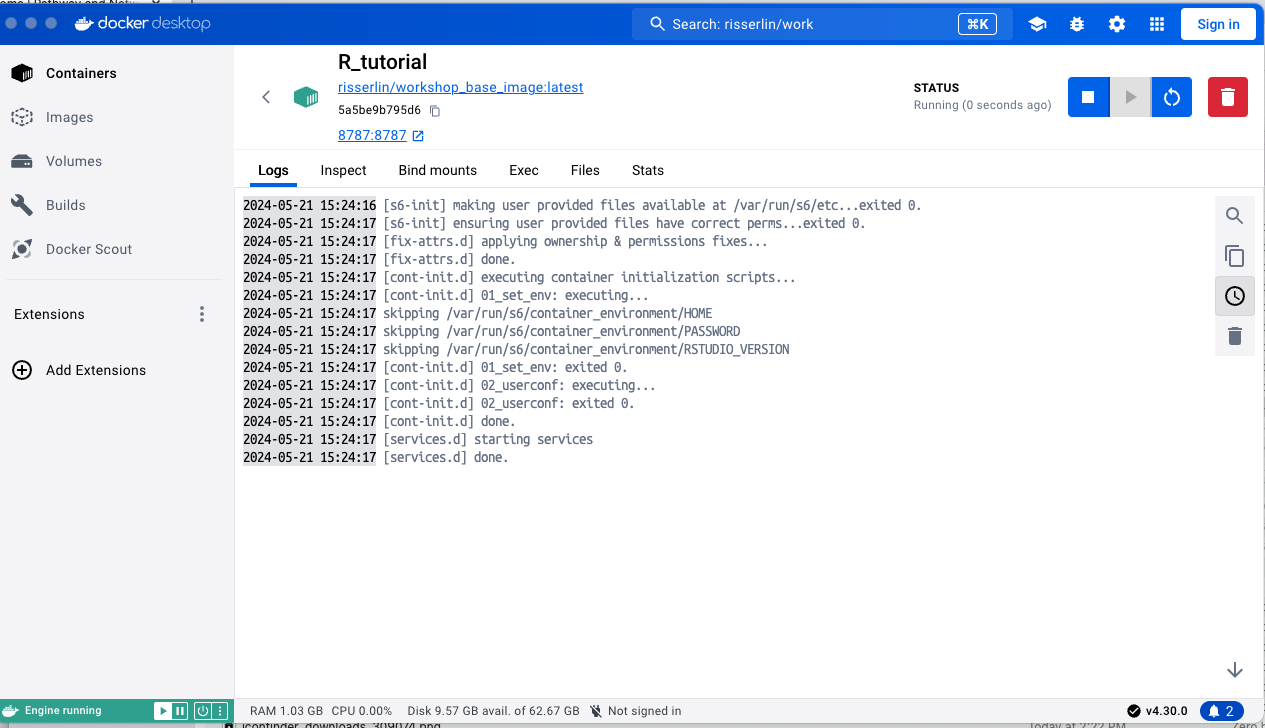
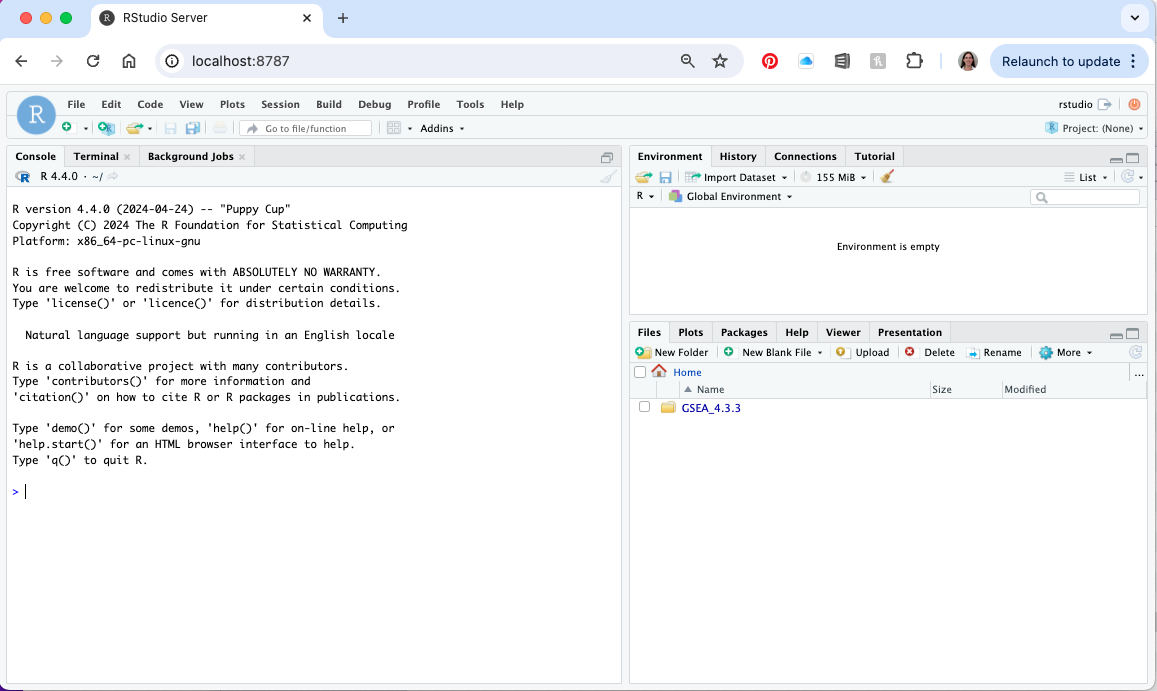
- Open a web browser and navigate to localhost:8787
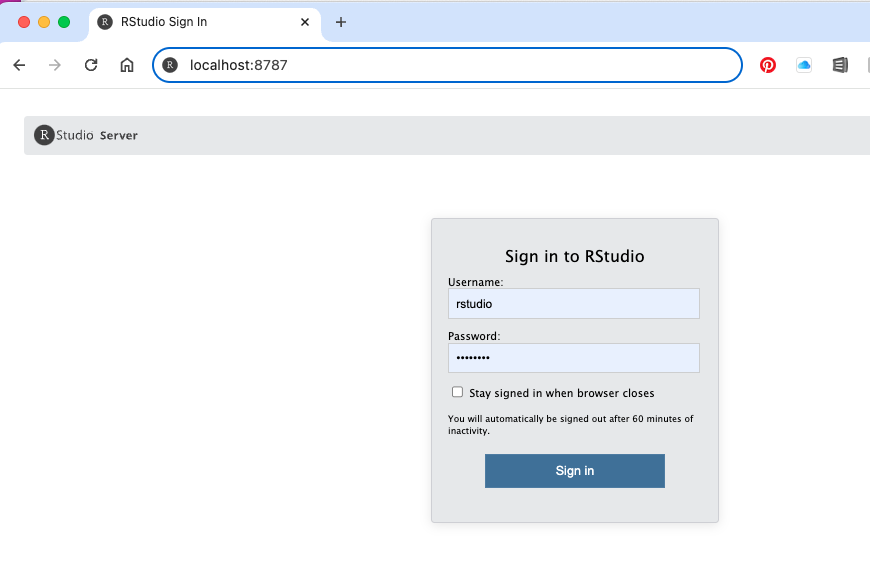
- Username - rstudio, password - password (or whatever you entered as the PASSWORD settting when you launch the container)
- You should now have an r studio session running in your web browser
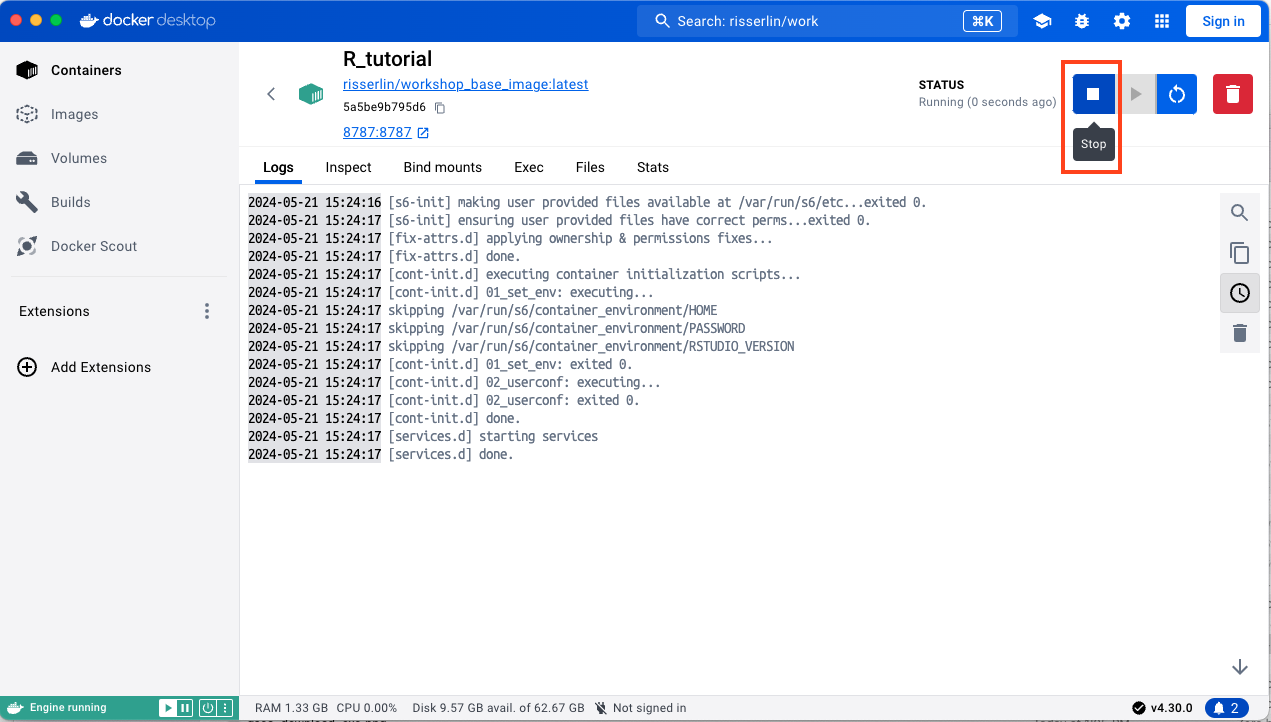
- When you are finished doing the tutorial remember to turn off your docker container and dacker as they both use up a lot of your computer’s resources.
Pre-workshop Readings and Lectures
- Video Module 1 - Introduction to Pathway and Network Analysis by Gary Bader
- Video Module 5 - Gene Function Prediction (GeneMania) by Quaid Morris
- Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, Merico D, Bader GD Nat Protoc. 2019 Feb;14(2):482-517 - Available here as well
Additional tutorials
iRegulon: from a gene list to a gene regulatory network using large motif and track collectionsJanky R, Verfaillie A, Imrichová H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, Svetlichnyy D, Kalender Atak Z, Fiers M, Marine JC, Aerts S PLoS Comput Biol. 2014 Jul 24;10(7)
The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, Maitland A, Mostafavi S, Montojo J, Shao Q, Wright G, Bader GD, Morris Q Nucleic Acids Res 2010 Jul 1;38 Suppl:W214-20 - Available here as well
GeneMANIA update 2018 Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, Morris Q Nucleic Acids Res. 2018 Jun 15 - Available here as well
How to visually interpret biological data using networks Merico D, Gfeller D, Bader GD Nature Biotechnology 2009 Oct 27, 921-924 - Available here as well
g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Reimand J, Kull M, Peterson H, Hansen J, Vilo J Nucleic Acids Res. 2007 Jul;35
g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J Nucleic Acids Res. 2019 May 8
Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP Proc Natl Acad Sci U S A. 2005 Oct 25;102(43)
Expression data analysis with Reactome Jupe S, Fabregat A, Hermjakob H Curr Protoc Bioinformatics. 2015 Mar 9;49:8.20.1-9
Interacting with Cytoscape using CyRest and command lines (for advanced users): https://github.com/cytoscape/cytoscape-automation/blob/master/for-scripters/R/advanced-cancer-networks-and-data-rcy3.Rmd