Enrichment Map Workflow
Ruth Isserlin
2023-03-22
Set of R notebooks to transform expression data to a ranked list and run them through Pathay enrichment pipeline. Pathway enrichment analysis helps gain mechanistic insight into large gene lists typically resulting from genome scale (–omics) experiments. It identifies biological pathways that are enriched in the gene list more than expected by chance. We explain pathway enrichment analysis and present a practical step-by-step guide to help interpret gene lists. The protocol is designed for biologists with no prior bioinformatics training and uses freely available software including g:Profiler, GSEA, Cytoscape and Enrichment Map.
The main protocol involves translating a ranked list to an Enrichment map. The figure below outlines the main steps.
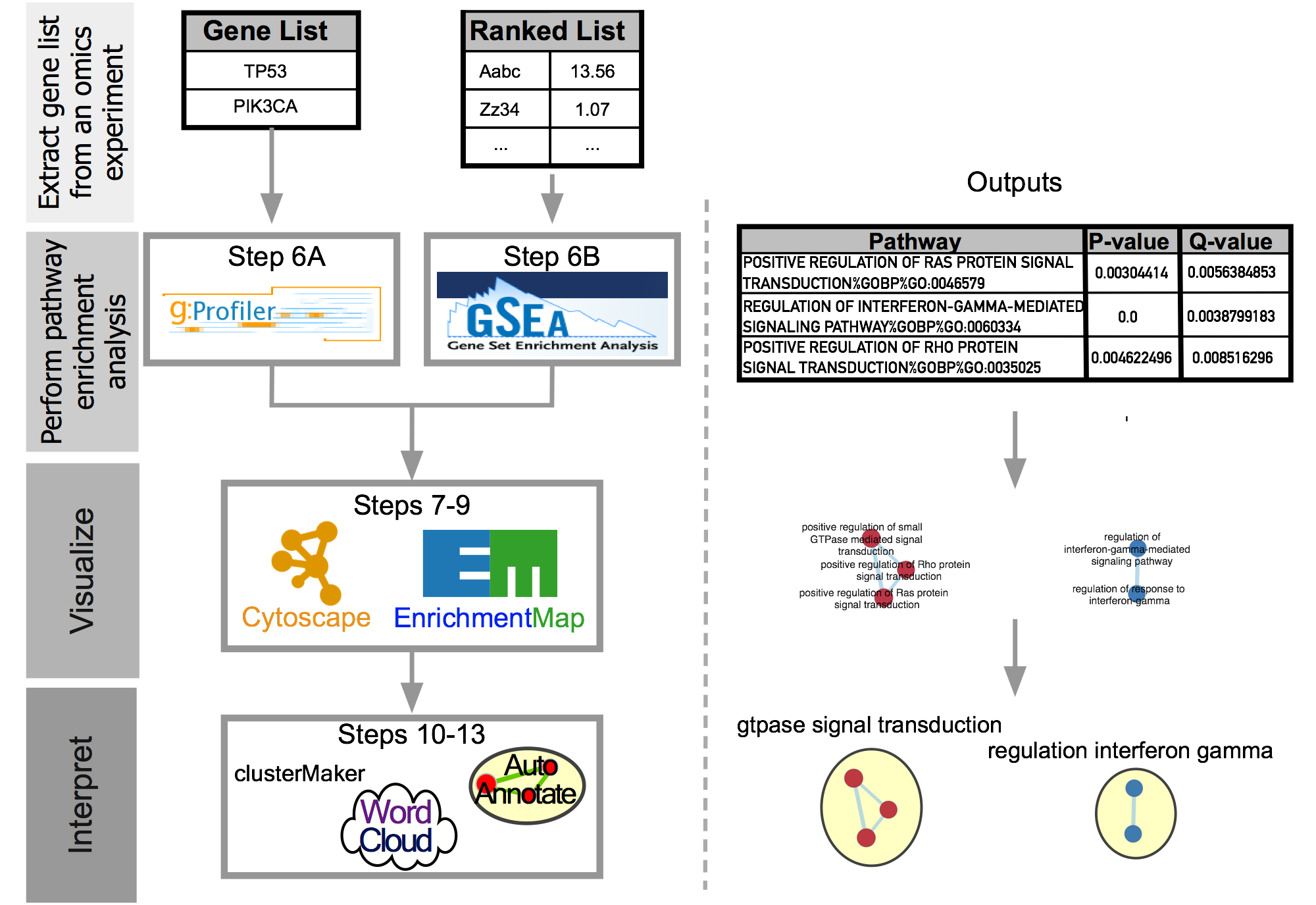
EM Protocol Flow Chart
Table of Contents
- Download TCGA data - R notebook shows you how to download legacy microarray and rnaseq ovarian cancer data. The notebook can be modified to download any data from GDC
- Supplemental Protocol 1 - convert raw RNASeq expression data to a ranked list
- Supplemental Protocol 2 - convert RMA normalized microarray expression data to a ranked list
- Supplemental Protocol 3 - perform Pathway Enrichment Analysis in R using ROAST and Camera
- Supplemental Protocol 4 - perform phenotype randomizations using edgeR with GSEA.
- Supplemental Protocol 5 - Creates a Multi dataset Enrichment Map (using GSEA results from multiple rank files) and followed by Theme calculation and summary.
- Protocol 2 - run GSEA on ranked list and automatically create an Enrichment Map from the results.
- Protocol 2 add on - take the resulting EM from Protocol 2 and annotate it. Pull all the annotations directly into R to be used for further analysis.
Data Files used for the Protocol
- Supplmentary table 1 - Cancer drivers. List of genes used as g:Profiler input.
- Supplmentary table 2 - GSEA Rank file. List of genes and their assoicated ranks used as input for GSEA.
- Supplmentary table 3 - GSEA GMT file. Pathway defintion file used for the protocol as GSEA input. Updated gmt files can be found on the baderlab download site here
- Supplmentary table 4 - Example g:Profiler results file.
- Supplmentary
table 5 - Example g:Profiler gmt file. If using example g:Profiler
results file instead of regenerating the results make sure to use this
gmt file. If the g:Profiler results file has been regenerated then make
sure to download an updated gmt file that corresponds to the results
file as specified in the protocol.
- Supplmentary
table 6 - Example expression file (corresponding to example rank
file, supplementary 2) used when creating an Enrichment Map from GSEA
results.
- Supplmentary table 7 - Example class file (corresponding to example expression file, supplementary 6) used when creating an Enrichment Map from GSEA results.
- Supplmentary table 8 - Example GSEA results file 1.
- Supplmentary table 9 - Example GSEA results file 2.
- Supplmentary table 10 - Example RANSeq expression input file used in Supplementary Protocol 1. File used to generate rank file, supplementary table 2, used in the main protocol.
- Supplmentary
table 11 - Example sample definition input file used in
Supplementary Protocol 1. File used to generate rank file, supplementary
table 2, used in the main protocol.
- Supplmentary
table 12 - Example Microarray expression input file used in
Supplementary Protocol 2.
- Supplmentary table 13 - Example sample definition input file used in Supplementary Protocol 2.
Download all the above files here
See the GitHub repository for the source.